Preface
In 1999, I packed everything I owned into my car for a cross-country trip to begin my new job as Staff Researcher at the University of California, Berkeley Computer Science Department. It was an optimistic time in my life and the country in general. The economy was well into the dot-com boom and still a few years away from the dot-com bust. Private investors were still happily throwing money at any company whose name started with an “e-” and ended with “.com”.
The National Science Foundation (NSF) was also funding ambitious digital projects like the National Partnership for Advanced Computing Infrastructure (NPACI). The goal of NPACI was to advance science by creating a pervasive national computational infrastructure called, at the time, “the Grid.” Berkeley was one of dozens of universities and affiliated government labs committed to connecting and sharing their computational and storage resources.
When I arrived at Berkeley, the Network of Workstations (NOW) project was just coming to a close. The NOW team had clustered together Sun workstations using Myrinet switches and specialized software to win RSA key-cracking challenges and break a number of sort benchmark records. The success of NOW led to a following project, the Millennium Project, that aimed to support even larger clusters built on x86 hardware and distributed across the Berkeley campus.
Ganglia exists today because of the generous support by the NSF for the NPACI project and the Millennium Project. Long-term investments in science and education benefit us all; in that spirit, all proceeds from the sales of this book will be donated to Scholarship America, a charity that to date has helped 1.7 million students follow their dreams of going to college.
Of course, the real story lies in the people behind the projects—people such as Berkeley Professor David Culler, who had the vision of building powerful clusters out of commodity hardware long before it was common industry practice. David Culler’s cluster research attracted talented graduated students, including Brent Chun and Matt Welsh, as well as world-class technical staff such as Eric Fraser and Albert Goto. Ganglia’s use of a lightweight multicast listen/announce protocol was influenced by Brent Chun’s early work building a scalable execution environment for clusters. Brent also helped me write an academic paper on Ganglia[1] and asked for only a case of Red Bull in return. I delivered. Matt Welsh is well known for his contributions to the Linux community and his expertise was invaluable to the broader teams and to me personally. Eric Fraser was the ideal Millennium team lead who was able to attend meetings, balance competing priorities, and keep the team focused while still somehow finding time to make significant technical contributions. It was during a “brainstorming” (pun intended) session that Eric came up with the name “Ganglia.” Albert Goto developed an automated installation system that made it easy to spin up large clusters with specific software profiles in minutes. His software allowed me to easily deploy and test Ganglia on large clusters and definitely contributed to the speed and quality of Ganglia development.
I consider myself very lucky to have worked with so many talented professors, students, and staff at Berkeley.
I spent five years at Berkeley, and my early work was split between NPACI and Millennium. Looking back, I see how that split contributed to the way I designed and implemented Ganglia. NPACI was Grid-oriented and focused on monitoring clusters scattered around the United States; Millennium was focused on scaling software to handle larger and larger clusters. The Ganglia Meta Daemon (gmetad)—with its hierarchical delegation model and TCP/XML data exchange—is ideal for Grids. I should mention here that Federico Sacerdoti was heavily involved in the implementation of gmetad and wrote a nice academic paper[2] highlighting the strength of its design. On the other hand, the Ganglia Monitoring Daemon (gmond)—with its lightweight messaging and UDP/XDR data exchange—is ideal for large clusters. The components of Ganglia complement each other to deliver a scalable monitoring system that can handle a variety of deployment scenarios.
In 2000, I open-sourced Ganglia and hosted the project from a Berkeley website. You can still see the original website today using the Internet Archive’s Wayback Machine. The first version of Ganglia, written completely in C, was released on January 9, 2001, as version 1.0-2. For fun, I just downloaded 1.0-2 and, with a little tweaking, was able to get it running inside a CentOS 5.8 VM on my laptop.
I’d like to take you on a quick tour of Ganglia as it existed over 11 years ago!
Ganglia 1.0-2 required you to deploy a daemon process, called a
dendrite, on every machine in your cluster. The
dendrite would send periodic heartbeats as well as publish any significant
/proc metric changes on a common multicast channel. To collect
the dendrite updates, you deployed a single instance of a
daemon process, called an axon, that indexed the metrics in
memory and answered queries from a command-line utility named
ganglia.
If you ran ganglia without any options, it would output
the following help:
$ ganglia
GANGLIA SYNTAX
ganglia [+,-]token [[+,-]token]...[[+,-]token] [number of nodes]
modifiers
+ sort ascending (default)
- sort descending
tokens
cpu_num cpu_speed cpu_user cpu_nice cpu_system
cpu_idle cpu_aidle load_one load_five load_fifteen
proc_run proc_total rexec_up ganglia_up mem_total
mem_free mem_shared mem_buffers mem_cached swap_total
swap_free
number of nodes
the default is all the nodes in the cluster or GANGLIA_MAX
environment variables
GANGLIA_MAX maximum number of hosts to return
(can be overidden by command line)
EXAMPLES
prompt> ganglia -cpu_num
would list all (or GANGLIA_MAX) nodes in ascending order by number of cpus
prompt> ganglia -cpu_num 10
would list 10 nodes in descending order by number of cpus
prompt> ganglia -cpu_user -mem_free 25
would list 25 nodes sorted by cpu user descending then by memory free ascending
(i.e., 25 machines with the least cpu user load and most memory available)As
you can see from the help page, the first version of ganglia
allowed you to query and sort by 21 different system metrics right out of
the box. Now you know why Ganglia metric names look so much like
command-line arguments (e.g., cpu_num, mem_total).
At one time, they were!
The output of the ganglia command made it very easy to
embed it inside of scripts. For example, the output from Example 1 could be used to autogenerate an MPI machine file
that contained the least-loaded machines in the cluster for load-balancing
MPI jobs. Ganglia also automatically removed hosts from the list that had
stopped sending heartbeats to keep from scheduling jobs on dead
machines.
$ ganglia -load_one 10 hpc0991 0.10 hpc0192 0.10 hpc0381 0.07 hpc0221 0.06 hpc0339 0.06 hpc0812 0.02 hpc0042 0.01 hpc0762 0.01 hpc0941 0.00 hpc0552 0.00
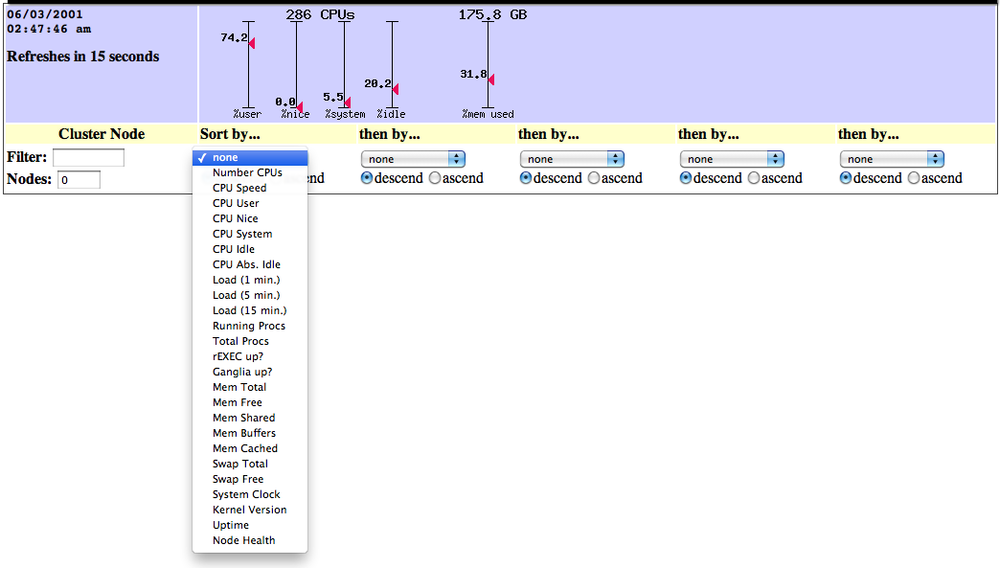
Ganglia 1.0-2 had a simple UI written in PHP 3 that would query an
axon and present the response as a dynamic graph of aggregate
cluster CPU and memory utilization as well as the requested metrics in
tabular format. The UI allowed for filtering by hostname and could limit the
total number of hosts displayed.
Ganglia has come a very long way in the last 11 years! As you read this book, you’ll see just how far the project has come.
Ganglia 1.0 ran only on Linux, whereas Ganglia today runs on dozens of platforms.
Ganglia 1.0 had no time-series support, whereas Ganglia today leverages the power of Tobi Oetiker’s RRDtool or Graphite to provide historical views of data at granularities from minutes to years.
Ganglia 1.0 had only a basic web interface, whereas Ganglia today has a rich web UI (see Figure 1) with customizable views, mobile support, live dashboards, and much more.
Ganglia 1.0 was not extensible, whereas Ganglia today can publish custom metrics via Python and C modules or a simple command-line tool.
Ganglia 1.0 could only be used for monitoring a single cluster, whereas Ganglia today can been used to monitor hundreds of clusters distributed around the globe.
I just checked our download stats and Ganglia has been downloaded more than 880,000 times from our core website. When you consider all the third-party sites that distribute Ganglia packages, I’m sure the overall downloads are well north of a million!
Although the NSF and Berkeley deserve credit for getting Ganglia started, it’s the generous support of the open source community that has made Ganglia what it is today. Over Ganglia’s history, we’ve had nearly 40 active committers and hundreds of people who have submitted patches and bug reports. The authors and contributors on this book are all core contributors and power users who’ll provide you with the in-depth information on the features they’ve either written themselves or use every day.
Reflecting on the history and success of Ganglia, I’m filled with a lot of pride and only a tiny bit of regret. I regret that it took us 11 years before we published a book about Ganglia! I’m confident that you will find this book is worth the wait. I’d like to thank Michael Loukides, Meghan Blanchette, and the awesome team at O’Reilly for making this book a reality.
—Matt Massie
Conventions Used in This Book
The following typographical conventions are used in this book:
- Italic
Indicates new terms, URLs, email addresses, filenames, and file extensions.
Constant widthUsed for program listings, as well as within paragraphs to refer to program elements such as variable or function names, databases, data types, environment variables, statements, and keywords.
Constant width boldShows commands or other text that should be typed literally by the user.
Constant width italicShows text that should be replaced with user-supplied values or by values determined by context.
Tip
This icon signifies a tip, suggestion, or general note.
Caution
This icon indicates a warning or caution.
Using Code Examples
This book is here to help you get your job done. In general, you may use the code in this book in your programs and documentation. You do not need to contact us for permission unless you’re reproducing a significant portion of the code. For example, writing a program that uses several chunks of code from this book does not require permission. Selling or distributing a CD-ROM of examples from O’Reilly books does require permission. Answering a question by citing this book and quoting example code does not require permission. Incorporating a significant amount of example code from this book into your product’s documentation does require permission.
We appreciate, but do not require, attribution. An attribution usually includes the title, author, publisher, and ISBN. For example: “Monitoring with Ganglia by Matt Massie, Bernard Li, Brad Nicholes, and Vladimir Vuksan (O’Reilly). Copyright 2013 Matthew Massie, Bernard Li, Brad Nicholes, Vladimir Vuksan, 978-1-449-32970-9.”
If you feel your use of code examples falls outside fair use or the permission given above, feel free to contact us at permissions@oreilly.com.
Safari® Books Online
Note
Safari Books Online (www.safaribooksonline.com) is an on-demand digital library that delivers expert content in both book and video form from the world’s leading authors in technology and business.
Technology professionals, software developers, web designers, and business and creative professionals use Safari Books Online as their primary resource for research, problem solving, learning, and certification training.
Safari Books Online offers a range of product mixes and pricing programs for organizations, government agencies, and individuals. Subscribers have access to thousands of books, training videos, and prepublication manuscripts in one fully searchable database from publishers like O’Reilly Media, Prentice Hall Professional, Addison-Wesley Professional, Microsoft Press, Sams, Que, Peachpit Press, Focal Press, Cisco Press, John Wiley & Sons, Syngress, Morgan Kaufmann, IBM Redbooks, Packt, Adobe Press, FT Press, Apress, Manning, New Riders, McGraw-Hill, Jones & Bartlett, Course Technology, and dozens more. For more information about Safari Books Online, please visit us online.
How to Contact Us
Please address comments and questions concerning this book to the publisher:
| O’Reilly Media, Inc. |
| 1005 Gravenstein Highway North |
| Sebastopol, CA 95472 |
| 800-998-9938 (in the United States or Canada) |
| 707-829-0515 (international or local) |
| 707-829-0104 (fax) |
We have a web page for this book, where we list errata, examples, and any additional information. You can access this page at http://oreil.ly/ganglia.
To comment or ask technical questions about this book, send email to bookquestions@oreilly.com.
For more information about our books, courses, conferences, and news, see our website at http://www.oreilly.com.
Find us on Facebook: http://facebook.com/oreilly
Follow us on Twitter: http://twitter.com/oreillymedia
Watch us on YouTube: http://www.youtube.com/oreillymedia
[1] Massie, Matthew, Brent Chun, and David Culler. The Ganglia Distributed Monitoring System: Design, Implementation, and Experience. Parallel Computing, 2004. 0167-8191.
[2] Sacerdoti, Federico, Mason Katz, Matthew Massie, and David Culler. Wide Area Cluster Monitoring with Ganglia. Cluster Computing, December 2003.
Get Monitoring with Ganglia now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.