9.3 Results
9.3.1 Information Synergy for Simulated Gene Pairs
Existing network-based approaches typically incorporate differential analysis or correlation analysis to identify the genes or gene pairs specific to phenotypes or diseases. In order to illustrate how information synergy analysis differs with differential analysis and correlation analysis, we simulated scenarios where the genes vary in differential expression and correlation, and calculated the information synergy for each of the gene pairs. Six representative scenarios were simulated, including (1) both genes differentially expressed between phenotypes, and correlated in both phenotypes; (2) both genes differentially expressed, and have no correlation with each phenotype; (3) both genes are not differentially expressed, but correlated in both phenotype; (4) both genes are not differentially expressed, and have no correlation with each phenotype; (5) both genes are not differentially expressed, but similarly correlated in each phenotype; (6) both genes are not differentially expressed, but differentially correlated between phenotypes. The joint expression patterns of these six scenarios are provided in Figure 9.1, see panels a–f, respectively.
Figure 9.1 Information synergy scores for simulated gene pairs.
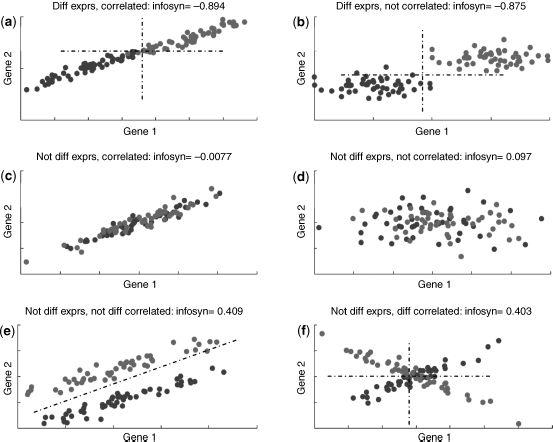
The information synergy scores for each simulated gene pair is also provided in Figure 9.1. The observations in ...
Get Statistical and Machine Learning Approaches for Network Analysis now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

