3.1 Introduction
The biomolecules of the living organisms, such as proteins and metabolites, undergo several interactions and chemical reactions, which lead to the occurrence of various life phenomena [1–6]. These interactions can be represented in the form of networks (graphs) (see Fig. 3.1, for example), the so-called “biological networks”; the evolution of these biological networks is a long-standing question. It is believed that biological networks adaptively shape-shift with the changing environment (e.g., temperature, pressure, and radial ray), and consequently, living organisms can perform new functions. Thus, for understanding life phenomena, it is important to obtain an understanding of network structures and their formation mechanisms from a macroscopic viewpoint. In addition to this, such a network approach also plays a significant role in technological processes such as finding missing interactions and designing novel interactions.
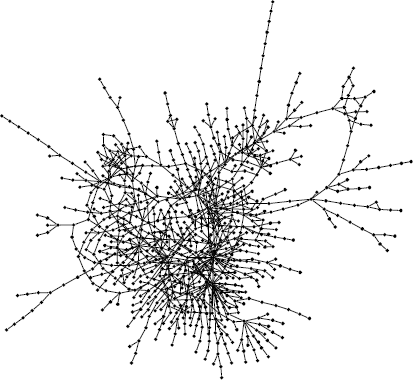
Figure 3.1 The partial metabolic network of Escherichia coli obtained from the KEGG database [7]. The nodes (filled circles) and edges (links) represent the metabolites and substrate–product relationships. This network was drawn using the yEd Graph Editor [8].
With recent developments in biotechnology and bioinformatics, the understanding of the interactions among biomolecules is progressively becoming clearer. The data of the interactions ...
Get Statistical and Machine Learning Approaches for Network Analysis now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

