7.4 Automated Morphological Endmember Extraction (AMEE)
The AMEE algorithm is an endmember extraction algorithm that makes simultaneous use of spatial and spectral information via multi-channel morphological processing (Plaza et al., 2002). The input to AMEE is the full image data cube, with no need of dimensionality reduction. Let r denote the input data cube and r(x,y) denote the pixel vector at spatial location (x,y). Similarly, let K be a kernel defined in the spatial domain of the image (the x–y plane). This kernel, usually called structuring element (SE) in mathematical morphology terminology, is translated over the image. The SE acts as a probe for extracting or suppressing specific structures of the image objects, according to the size and shape of the SE. Having the above definitions in mind, AMEE method is based on the application of multichannel erosion and dilation operations to the data. The above operations are defined as follows:
(7.22) 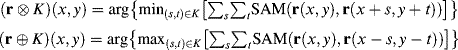
where SAM is the spectral angle mapper (SAM). Multichannel erosion (respectively, dilation) selects the pixel vector that minimizes (respectively, maximizes) a cumulative distance-based cost function, based on the sum of the SAM distance scores between each pixel in the spatial neighborhood and all the other pixels in the neighborhood. As a result, multichannel erosion extracts the pixel vector that is more similar to its neighbors ...
Get Hyperspectral Data Processing: Algorithm Design and Analysis now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

