Let's take a look at the following steps:
- We will start by inspecting the description of all of the the sequences on the reference genome FASTA file:
from Bio import SeqIOgenome_name = 'PlasmoDB-9.3_Pfalciparum3D7_Genome.fasta'recs = SeqIO.parse(genome_name, 'fasta')for rec in recs: print(rec.description)
This code should look familiar from the previous chapter, Chapter 2, Next-Generation Sequencing. Let's take a look at part of the output:
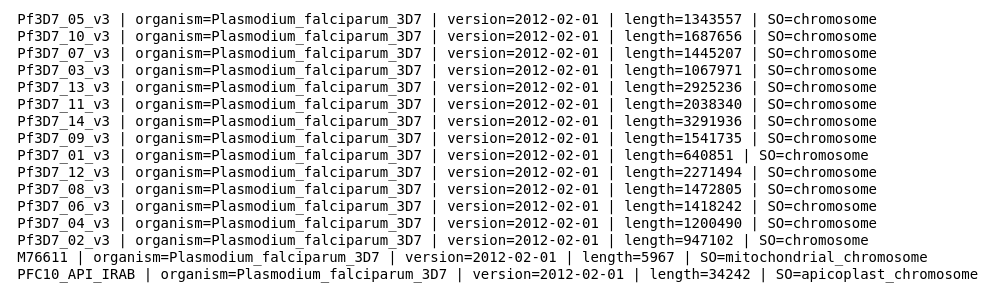
Different genome references will have different description lines, but they will generally have important information. In this example, you can see that we have chromosomes, mitochondria, ...

