CHAPTER 15tBLASTn
CS Mukhopadhyay and RK Choudhary
School of Animal Biotechnology, GADVASU, Ludhiana
15.1 INTRODUCTION
tBLASTn is another type of translated BLAST algorithm, in which an amino acid sequence is used as a query to compare with the translated nucleotide (coding sequence) database. The amino acid sequence is compared at the protein level with each subject nucleotide sequence translated in all six reading frames. Thus, tBLASTn is very useful for searching protein homolog(s) in unannotated nucleotide data such as expressed sequence tags (maintained in BLAST database “est”) and draft genome records (located in the BLAST database “htgs”), which remain unannotated in the respective databases.
15.2 OBJECTIVE
To search for the homologous protein sequences of a pair of given protein sequences (NP_001028007, NP_001028008).
15.3 PROCEDURE
The basic steps of tBLASTn are the same as for BLASTx:
15.3.1 Open the tBLASTn page
Open the NCBI home page by typing http://www.ncbi.nlm.nih.gov/ and click “tBLASTn”. Alternatively, it can also be opened by entering http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM =tblastn&PAGE_TYPE = BlastSearch&LINK_LOC = blasthome in the URL.
The main page of tBLASTn will be displayed (Figure 15.1).
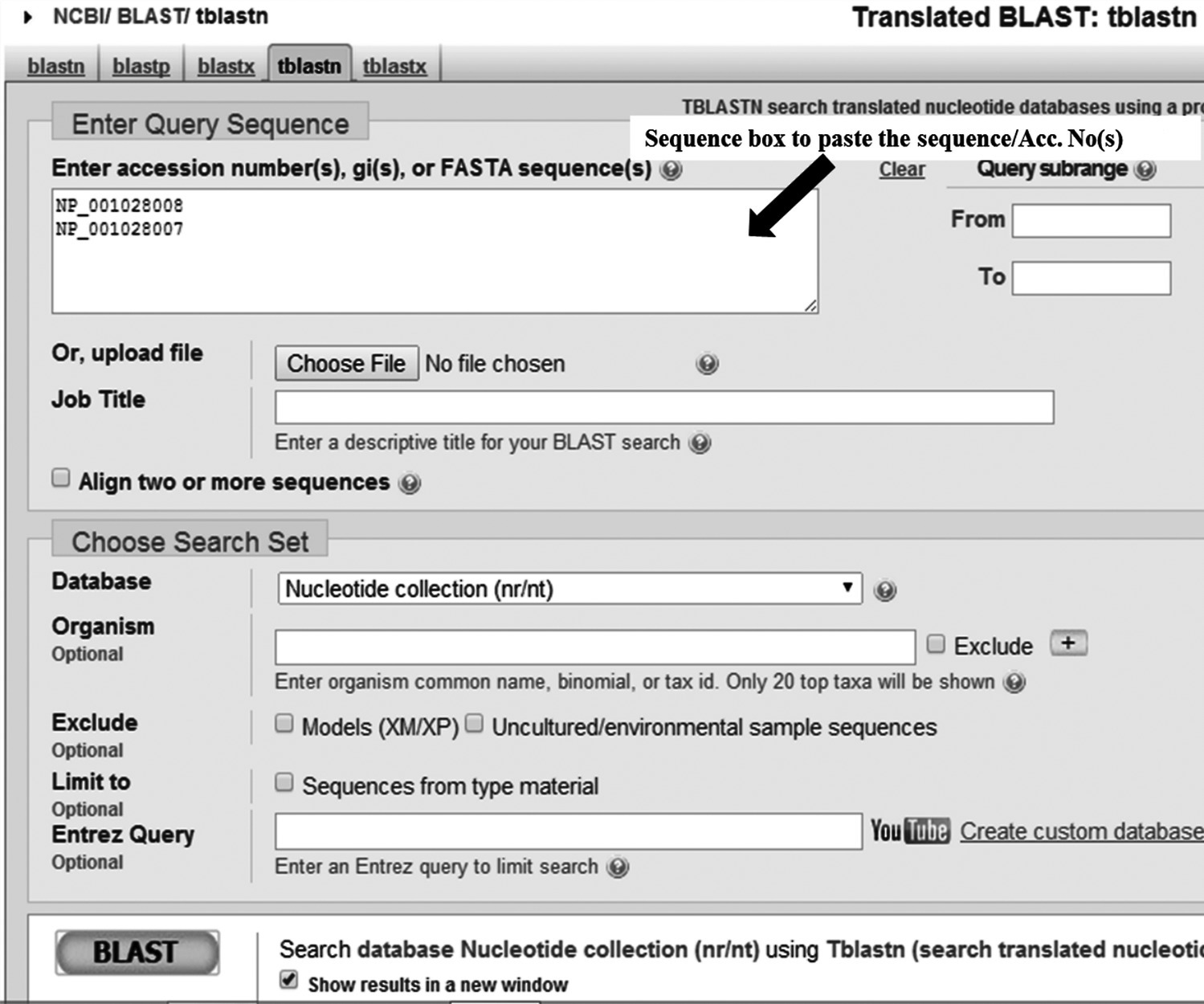
FIGURE 15.1 Homepage for tBLASTn at NCBI. The query sequence(s) can be entered with either accession numbers or sequence(s) in FASTA format.
15.3.2 ...
Get Basic Applied Bioinformatics now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

