This is the Title of the Book, eMatter Edition
Copyright © 2012 O’Reilly & Associates, Inc. All rights reserved.
14
|
Chapter 1: Hello BLAST
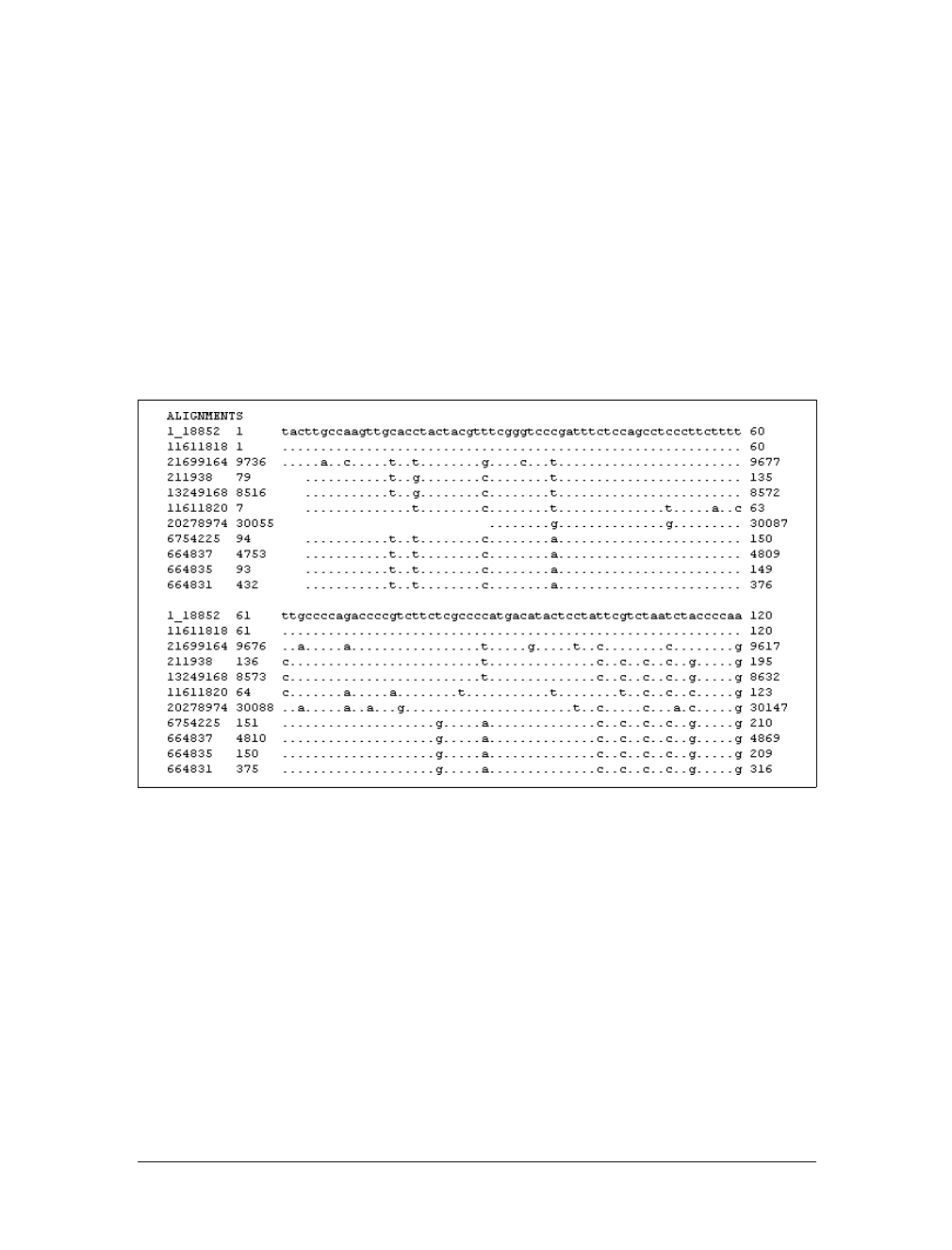
The other available alignment options allow a multiple sequence alignment view of
the BLAST hits. One of these multiple alignment options, query-anchored with iden-
tities, is shown in Figure 1-14. In this view, the full sequence of the query is shown
on the top line with a unique identifier (1_18852, in this case). Subsequently, each
line shows the alignment for one database hit. Identical residues are represented with
a dot (.), while nucleotide differences are shown explicitly. This alignment option is
useful for quickly identifying changes common to a group of sequences. For exam-
ple, you can see from the part of the alignment shown in Figure 1-14 that the bot-
tom four sequences (6754225, 664837, 664835, and 664831) have common shared
differences. A deeper look into these sequences reveals that they are actually differ-
ent database entries for the same mouse Hoxa11 gene, which is homologous to the
coelacanth Hoxa11 gene.
The other multiple sequence alignment views are similar to this one, but differ on
whether or not they show identical residues (with or without identities) and whether
the gaps are displayed in the query sequence or in the subjects (flat or not). You’ll
find a detailed explanation of these alignment options in Appendix A.
The Next Step
This chapter has taken you through a simple BLASTN search at the NCBI database;
however, more than two dozen specialized BLAST pages are available, and they let
you do anything—from screening for vector sequence, to identifying protein family
Figure 1-14. Query-anchored with identities view
Get BLAST now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

