Chapter 4. Control Statements
Note
This chapter’s material is rich and intellectually challenging. Don’t give up if you start to feel lost (but do review it later to make sure you have absorbed it all). This chapter, together with the next, will complete our introduction to Python. To help you understand its contents, the chapter ends with some extended examples that reiterate the points made in its shorter examples. The rest of the book has a very different flavor.
Chapters 1 and 2 introduced simple statements:
Expressions, including function calls
Assignments
Augmented assignments
Various forms of import
Assertions
returnyield(to implement generators)pass
They also introduced the statements def, for defining functions, and with, to use with files.[20] These are compound statements because they require at least one indented statement after the
first line. This chapter introduces other compound statements. As with
def and with, the first line of every compound statement
must end with a colon and be followed by at least one statement indented
relative to it. Unlike def and with statements, though, the other compound
statements do not name anything. Rather, they determine the order in which
other statements are executed. That order is traditionally called the
control flow or flow of control,
and statements that affect it are called control statements.[21]
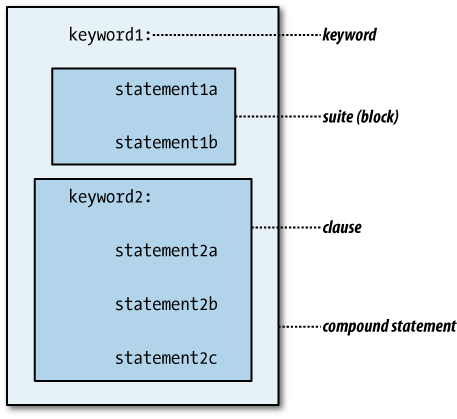
Some kinds of compound statements can or must have more than one clause. The first line of each clause of a compound statement—its header in Python terminology—is at the same level of indentation as the headers of the statement’s other clauses. Each header begins with a keyword and ends with a colon. The rest of the clause—its suite—is a series of statements indented one level more than its header.
Note
The term “suite” comes from Python’s technical documentation. We’ll
generally use the more common term block instead.
Also, when discussing compound statements, we frequently refer to clauses
by the keywords that introduce them (for example, “a with clause”).
Figure 4-1 illustrates
the structure of a multi-clause compound statement. Not all compound
statements are multi-clause, but every clause has a header and a suite
containing at least one statement (if only pass).
The statements discussed in this chapter are the scaffolding on which you will build your programs. Without them you are limited to the sort of simple computations shown in the examples of the previous chapters. The four kinds of compound statements introduced here are:
Conditionals
Loops
Iterations
Exception handlers
Note
Starting later in this chapter, some common usage patterns of Python functions, methods, and statements will be presented as abstract templates, along with examples that illustrate them. These templates are a device for demonstrating and summarizing how these constructs work while avoiding a lot of written description. They contain a mixture of Python names, self-descriptive “roles” to be replaced by real code, and occasionally some “pseudocode” that describes a part of the template in English rather than actual Python code.
The templates are in no way part of the Python language. In addition to introducing new programming constructs and techniques as you read, the templates are designed to serve as references while you work on later parts of the book and program in Python afterwards. Some of them are quite sophisticated, so it would be worth reviewing them periodically.
Conditionals
The most direct way to affect the flow of control is with a
conditional statement. Conditionals in Python are
compound statements beginning with if.
During the import of a module __name__ is bound to the name of the module, but
while the file is being executed __name__ is bound to '__main__'. This gives you a way to include
statements in your Python files that are executed only when the module is
run or, conversely, only when it is imported. The comparison of __name__ to '__main__' would almost always be done in a
conditional statement and placed at the end of the file.
A common use for this comparison is to run tests when the module is executed, but not when it is
imported for use by other code. Suppose you have a function called
do_tests that contains lots of
assignment and assertion statements that you don’t want to run when the
module is imported in normal use, but you do want to execute when the
module is executed. While informal, this is a useful technique for testing
modules you write. At the end of the file you would write:
if __name__ == '__main__':
do_tests()There are several forms of conditional statements. The next one
expresses a choice between two groups of statements and has two clauses,
an if and an else.
A simple use of the one-alternative form of conditional is to expand the test for whether a file is being imported as opposed to being executed. We can set it up so that one thing happens when the file is imported and a different thing happens when it’s executed.
This example shows only one statement in each block. There could be
others, but another reason to group statements into simple functions is so
you can invoke them “manually” in the interpreter during development and
testing. You might run do_tests, fix a
few things, then run it again. These test functions are useful whether
invoked automatically or manually:
if __name__ == '__main__':
do_tests()
else:
print(__name__, 'has been imported.')The third form of conditional statement contains more than one test.
Except for the if at the beginning, all
the test clauses are introduced by the keyword elif.
Python has a rich repertoire of mechanisms for controlling execution. Many kinds of maneuvers that would have been handled in older languages by conditionals—and could still be in Python—are better expressed using these other mechanisms. In particular, because they emphasize values rather than actions, conditional expressions or conditional comprehensions are often more appropriate than conditional statements.
Programming languages have many of the same properties as ordinary human languages. Criteria for clear writing are similar in both types of language. You want what you write to be:
Succinct
Clear
Accurate
Note
It’s important not to burden readers of your code (you included!) with too many details. People can pay attention to only a few things at once. Conditionals are a rather heavy-handed form of code that puts significant cognitive strain on the reader. With a little experience and experimentation, you should find that you don’t often need them. There will be examples of appropriate uses in the rest of this chapter, as well as in later ones. You should observe and absorb the style they suggest.
Loops
A loop is a block of statements that gets executed as long as some condition is
true. Loops are expressed in Python using while statements.
Note that the test may well be false the first time it is evaluated.
In that case, the statements of the block won’t get executed at all. If
you want some code to execute once the test is false, include an else clause in your loop.
There are two simple statements that are associated with both loops
and iterations (the subject of the next section): continue and break.
The continue statement is rarely used in Python programming, but it’s worth
mentioning it here in case you run across it while reading someone else’s
Python code. The break statement is
seen somewhat more often, but in most cases it is better to embed in the
loop’s test all the conditions that determine whether it should continue
rather than using break. Furthermore, in
many cases the loop is the last statement of a function, so you can just use a return statement to both end the loop and exit
the function. (A return exits the
function that contains it even if execution is in the middle of a
conditional or loop.) Using a return
instead of break is more convenient
when each function does just one thing: most uses of break are intended to move past the loop to
execute code that appears later in the function, and if there isn’t any
code later in the function a return
statement inside the loop is equivalent to a break.
An error that occurs during the execution of a loop’s test or one of its statements also terminates the execution of the loop. Altogether, then, there are three ways for a loop’s execution to end:
- Normally
The test evaluates to false.
- Abnormally
An error occurs in the evaluation of the test or body of the loop.
- Prematurely
The body of the loop executes a
returnorbreakstatement.
When you write a loop, you must make sure that the test expression
eventually becomes false or a break or
return is executed. Otherwise, the
program will get stuck in what is called an infinite loop. We’ll see at the end of
the chapter how to control what happens when errors occur, rather than
allowing them to cause the program to exit abnormally.
Simple Loop Examples
Example 4-1 presents the simplest possible loop, along with a function that reads a line typed by the user, prints it out, and returns it.
def echo(): """Echo the user's input until an empty line is entered""" while echo1(): pass def echo1(): """Prompt the user for a string, "echo" it, and return it""" line = input('Say something: ') print('You said', line) return line
The function echo1 reads a
line, prints it, and returns it. The function echo contains the simplest possible while statement. It calls a function
repeatedly, doing nothing (pass),
until the function returns something false. If the user just presses
Return, echo1 will print and return
an empty string. Since empty strings are false, when the while gets an empty string back from echo1 it stops. A slight variation, shown in
Example 4-2, is to compare the result returned from
echo1 to some specified value that
signals the end of the conversation.
def polite_echo(): """Echo the user's input until it equals 'bye'""" while echo1() != 'bye': pass
Of course, the bodies of loops are rarely so trivial. What allows
the loop in these examples to contain nothing but a pass is that echo1 is called both to perform an action and
to return True or False. This example uses such trivial loop
bodies only to illustrate the structure of the while statement.
Initialization of Loop Values
Example 4-3 shows a more typical loop. It records the user’s responses, and
when the user types 'bye' the
function returns a record of the input it received. The important thing
here is that it’s not enough to use echo1’s result as a test. The function also
needs to add it to a list it is building. That list is returned from the
function after the loop exits.
def recording_echo(): """Echo the user's input until it equals 'bye', then return a list of all the inputs received""" lst = [] entry = echo1() while entry != 'bye': lst.append(entry) entry = echo1() return lst
In this example, echo1 is
called in two places: once to get the first response and then each time
around the loop. Normally it is better not to repeat a piece of code in
two places, even if they are so close together. It’s easy to forget to
change one when you change the other or to make incompatible changes,
and changing the same thing in multiple places is tedious and
error-prone. Unfortunately, the kind of repetition shown in this example
is often difficult to avoid when combining input—whether from the user or from a file—with while loops.
Example 4-4 shows the same function as Example 4-3, but with comments added to emphasize the way the code uses a simple loop.
def recording_echo(): # initialize entry and lst lst = [] # get the first input entry = echo1() # test entry while entry != 'bye': # use entry lst.append(entry) # change entry entry = echo1() # repeat # return result return lst
All parts of this template are optional except for the line
beginning with while. Typically, one
or more of the values assigned in the initialization portion are used in
the loop test and changed inside the loop. In recording_echo the value of entry is initialized, tested, used, and
changed; lst is initialized, used,
and changed, but it is not part of the loop’s test.
Looping Forever
Sometimes you just want your code to repeat something until it executes a return statement. In that case there’s no need
to actually test a value. Since while
statements require a test, we use True, which is, of course, always true. This
may seem a bit odd, but there are times when something like this is
appropriate. It is often called “looping forever.” Of course, in reality
the program won’t run “forever,” but it might run forever as far as
it is concerned—that is, until something external
causes it to stop. Such programs are found frequently in operating system and server
software.
Example 4-5 shows a rewrite of Example 4-3 using the Loop Forever template. Typical loops usually get the next value at the end of the loop, but in this kind, the next value is obtained at the beginning of the loop.
def recording_echo_with_conditional(): """Echo the user's input until it equals 'bye', then return a list of all the inputs received""" seq = [] # no need to initialize a value to be tested since nothing is tested! while True: entry = echo1() if entry == 'bye': return seq seq.append(entry)
Loops over generators are always effectively “forever” in that
there’s no way to know how many items the generator will produce. The
program must call next over and over
again until the generator is exhausted. We saw in Chapter 3 (in Generators) that the
generator argument of next can be
followed by a value to return when the generator is exhausted. A
“forever” loop can be written to use this feature in a function that
combines all of the generated amino acid abbreviations into a string.
Example 4-6 repeats the
definition of the generator function and shows the definition of a new
function that uses it.
def aa_generator(rnaseq): """Return a generator object that produces an amino acid by translating the next three characters of rnaseq each time next is called on it""" return (translate_RNA_codon(rnaseq[n:n+3]) for n in range(0, len(rnaseq), 3)) def translate(rnaseq): """Translate rnaseq into amino acid symbols""" gen = aa_generator(rnaseq) seq = '' aa = next(gen, None) while aa: seq += aa aa = next(gen, None) return seq
Loops with Guard Conditions
Loops are often used to search for a value that meets the test condition when there is no guarantee that one does. In such situations it is not enough to just test each value—when there are no more values, the test would be repeated indefinitely. A second conditional expression must be added to detect that no more values remain to be tested.
Loops like these are used when there are two separate reasons for
them to end: either there are no more values to
use—at-end—or some kind of special value has
been encountered, detected by at-target. If
there are no more values to consider, evaluating
at-target would be meaningless, or, as is
often the case, would cause an error. The and operator is used to “protect” the second part of the test so that
it is evaluated only if the first is true. This is sometimes called
a guard condition.
When a loop can end for more than one reason, the statements after
the while will need to distinguish
the different cases. The simplest and most common case is to return one
value if the loop ended because at-end became
true and a different value if the loop ended because
at-target became true.
Two-condition loops like this occur frequently in code that reads from streams such as files, terminal input, network connections, and so on. That’s because the code cannot know when it has reached the end of the data until it tries to read past it. Before the result of a read can be used, the code must check that something was actually read.
Because readline returns
'\n' when it reads a blank line but
returns '' at the end of a file, it
is sufficient to check to see that it returned a nonempty string.
Repeated calls to readline at the end
of the file will continue to return empty strings, so if its return
value is not tested the loop will never terminate.
Example 4-7 shows a
function that reads the first sequence from a FASTA file. Each time it reads a line it must check first
that the line is not empty, indicating that the end of file has been
reached, and if not, that the line does not begin with '>', indicating the beginning of the next
sequence.
def read_sequence(filename): """Given the name of a FASTA file named filename, read and return its first sequence, ignoring the sequence's description""" seq = '' with open(filename) as file: line = file.readline() while line and line[0] == '>': line = file.readline() while line and line[0] != '>': # must check for end of file seq += line line = file.readline() return seq
Note
Although files can often be treated as collections of lines
using comprehensions or readlines,
in some situations it is more appropriate to loop using readline. This is especially true when
several related functions all read from the same stream.
The bare outline of code that loops over the lines of a file, doing something to each, is shown in the next template.
Iterations
Collections contain objects; that’s more or less all they
do. Some built-in functions—min, max, any, and
all—work for any type of collection.
The operators in and not in accept any type of collection as their
second operand. These functions and operators have something very
important in common: they are based on doing something with each element
of the collection.[22] Since any element of a collection could be its minimum or
maximum value, min and max must consider all the elements. The
operators in and not in and the functions any and all
can stop as soon as they find an element that meets a certain condition,
but if none do they too will end up considering every element of the
collection.
Doing something to each element of a collection is called iteration. We’ve actually already seen
a form of iteration—comprehensions. Comprehensions “do something” to every
element of a collection, collecting the results of that “something” into a
set, list, or dictionary. Comprehensions with multiple for clauses perform
nested iterations. Comprehensions with one or more if
clauses perform conditional iteration: unless an element passes all the
if tests, the “something” will not be
performed for that element and no result will be added.
The code in this book uses comprehensions much more aggressively
than many Python programmers do. You should get comfortable using them,
because in applicable situations they say just what they mean and say it
concisely. Their syntax emphasizes the actions and tests performed on the
elements. They produce collection objects, so the result of a
comprehension can be used in another expression, function call, return statement, etc. Comprehensions help
reduce the littering of code with assignment statements and the names they
bind.
The question, then, is: what kinds of collection manipulations do
not fit the mold of Python’s comprehensions? From the
point of view of Python’s language constructs, the answer is that actions
performed on each element of a collection sometimes must be expressed
using statements, and comprehensions allow only expressions.
Comprehensions also can’t stop before the end of the collection has been
reached, as when a target value has been located. For these and other
reasons Python provides the for
statement to perform general-purpose iteration over collections.
Iteration Statements
Iteration statements all begin with the keyword for. This section shows many ways for statements can be used and “templates”
that summarize the most important.
You will use for statements
often, since so much of your programming will use collections. The
for statement makes its purpose very
clear. It is easy to read and write and minimizes the opportunities for
making mistakes. Most importantly, it works for collections that aren’t
sequences, such as sets and dictionaries. As a matter of fact, the
for statement isn’t even restricted
to collections: it works with objects of a broader range of types that
together are categorized as iterables. (For
instance, we are treating file objects as collections (streams), but technically they are another
kind of iterable.)
Note
The continue and break statements introduced in the section
on loops work for iterations too.
By default, a dictionary iteration uses the dictionary’s keys. If
you want the iteration to use its values, call the values method
explicitly. To iterate with both keys and values at the same time, call
the items method. Typically, when
using both keys and values you would unpack the result of items and assign a
name to each, as shown at the end of the following template.
The previous chapter pointed out that if you need a dictionary’s
keys, values, or items as a list you can call list on the result of the corresponding
method. This isn’t necessary in for
statements—the results of the dictionary methods can be used
directly. keys, values, and items each return an iterable of a different
type—dict_keys, dict_values, and dict_items, respectively—but this difference
almost never matters, since the results of calls to these methods are
most frequently used in for
statements and as arguments to list.
Sometimes it is useful to generate a sequence of integers along
with the values over which a for
statement iterates. The function enumerate(iterable) generates tuples of the form (n, value), with n starting
at 0 and incremented with each value
taken from the iterable. It is rarely used anywhere but in a for statement.
A common use for enumerate is
to print out the elements of a collection along with a sequence of
corresponding numbers. The “do something” line of the template becomes a
call to print like the following:
print(n, value, sep='\t')
Kinds of Iterations
Most iterations conform to one of a small number of patterns. Templates for these patterns and examples of their use occupy most of the rest of this chapter. Many of these iteration patterns have correlates that use loops. For instance, Example 4-3 is just like the Collect template described shortly. In fact, anything an iteration can do can be done with loops. In Python programming, however, loops are used primarily to deal with external events and to process files by methods other than reading them line by line.
Note
Iteration should always be preferred over looping. Iterations are a clearer and more concise way to express computations that use the elements of collections, including streams. Writing an iteration is less error-prone than writing an equivalent loop, because there are more details to “get right” in coding a loop than in an iteration.
Do
Often, you just want to do something to every element of a collection. Sometimes that means calling a function and ignoring its results, and sometimes it means using the element in one or more statements (since statements don’t have results, there’s nothing to ignore).
A very useful function to have is one that prints every element
of a collection. When something you type into the interpreter returns
a long collection, the output is usually difficult to read.
Using pprint.pprint helps,
but for simple situations the solution demonstrated in Example 4-8 suffices. Both
pprint and this definition can be
used in other code too, of course.
def print_collection(collection): for item in collection: print(item) print()
Actually, even this action could be expressed as a comprehension:
[print(item) for item in collection]
Since print returns None, what you’d get
with that comprehension is a list containing one None value for each item in the collection.
It’s unlikely that you’d be printing a list large enough for it to
matter whether you constructed another one, but in another situation
you might call some other no-result function for a very large
collection. However, that would be silly and inefficient. What you
could do in that case is use a set, instead of list,
comprehension:
{print(item)for item in collection}That way the result would simply be {None}. Really, though, the use of a
comprehension instead of a Do iteration is not a serious suggestion,
just an illustration of the close connection between comprehensions
and iterations.
We can create a generalized “do” function by passing the “something” as a functional argument, as shown in Example 4-9.
The function argument could be a named function. For instance,
we could use do to redefine
print_collection from Example 4-8, as shown in
Example 4-10.
def print_collection(collection): do(collection, print)
This passes a named function as the argument to do. For more ad hoc uses we could pass a
lambda expression, as in Example 4-11.
The way to express a fixed number of repetitions of a block of code is to iterate over a range, as shown in the following template.
Collect
Iterations often collect the results of the “something” that gets done for each element. That means creating a new list, dictionary, or set for the purpose and adding the results to it as they are computed.
Most situations in which iteration would be used to collect results would be better expressed as comprehensions. Sometimes, though, it can be tricky to program around the limitation that comprehensions cannot contain statements. In these cases, a Collect iteration may be more straightforward. Perhaps the most common reason to use a Collect iteration in place of a comprehension or loop is when one or more names are assigned and used as part of the computation. Even in those cases, it’s usually better to extract that part of the function definition and make it a separate function, after which a call to that function can be used inside a comprehension instead of an iteration.
Example 4-12 shows a rewrite of the functions for reading entries from FASTA files in Chapter 3. In the earlier versions, all the entries were read from the file and then put through a number of transformations. This version, an example of the Collect iteration template, reads each item and performs all the necessary transformations on it before adding it to the collection. For convenience, this example also repeats the most succinct and complete comprehension-based definition.
While more succinct, and therefore usually more appropriate for most situations, the comprehension-based version creates several complete lists as it transforms the items. Thus, with a very large FASTA file the comprehension-based version might take a lot more time or memory to execute. After the comprehension-based version is yet another, this one using a loop instead of an iteration. You can see that it is essentially the same, except that it has extra code to read the lines and check for the end of the file.
def read_FASTA_iteration(filename): sequences = [] descr = None with open(filename) as file: for line in file: if line[0] == '>': if descr: # have we found one yet? sequences.append((descr, seq)) descr = line[1:-1].split('|') seq = '' # start a new sequence else: seq += line[:-1] sequences.append((descr, seq)) # add the last one found return sequences def read_FASTA(filename): with open(filename) as file: return [(part[0].split('|'), part[2].replace('\n', '')) for part in [entry.partition('\n') for entry in file.read().split('>')[1:]]] def read_FASTA_loop(filename): sequences = [] descr = None with open(filename) as file: line = file.readline()[:-1] # always trim newline while line: if line[0] == '>': if descr: # any sequence found yet? sequences.append((descr, seq)) descr = line[1:].split('|') seq = '' # start a new sequence else: seq += line line = file.readline()[:-1] sequences.append((descr, seq)) # easy to forget! return sequences
Combine
Sometimes we want to perform an operation on all of the elements of a collection to
yield a single value. An important feature of this kind of iteration
is that it must begin with an initial value. Python has a
built-in sum function but no
built-in product; Example 4-13 defines one.
def product(coll): """Return the product of the elements of coll converted to floats, including elements that are string representations of numbers; if coll has an element that is a string but doesn't represent a number, an error will occur""" result = 1.0 # initialize for elt in coll: result *= float(elt) # combine element with return result # accumulated result
As simple as this definition is, there is no reasonable way to define it just using a comprehension. A comprehension always creates a collection—a set, list, or dictionary—and what is needed here is a single value. This is called a Combine (or, more technically, a “Reduce”[23]) because it starts with a collection and ends up with a single value.
For another example, let’s find the longest sequence in a
FASTA file. We’ll assume we have a function called
read_FASTA, like one of the
implementations shown in Chapter 3. Example 4-13 used a binary operation to
combine each element with the previous result. Example 4-14 uses a
two-valued function instead, but the idea is the same. The inclusion
of an assignment statement inside the loop is an indication that the
code is doing something that cannot be done with a
comprehension.
def longest_sequence(filename): longest_seq = '' for info, seq in read_FASTA(filename): longest_seq = max(longest_seq, seq, key=len) return longest_seq
A special highly reduced form of Combine is Count, where all the iteration does is count the number of elements. It would be used to count the elements in an iterable that doesn’t support length. This template applies particularly to generators: for a generator that produces a large number of items, this is far more efficient than converting it to a list and then getting the length of the list.
One of the most important and frequently occurring kinds of actions on iterables that cannot be expressed as a comprehension is one in which the result of doing something to each element is itself a collection (a list, usually), and the final result is a combination of those results. An ordinary Combine operation “reduces” a collection to a value; a Collection Combine reduces a collection of collections to a single collection. (In the template presented here the reduction is done step by step, but it could also be done by first assembling the entire collection of collections and then reducing them to a single collection.)
Example 4-15 shows an example in which “GenInfo” IDs are extracted from each of several files, and a single list of all the IDs found is returned.
def extract_gi_id(description): """Given a FASTA file description line, return its GenInfo ID if it has one""" if line[0] != '>': return None fields = description[1:].split('|') if 'gi' not in fields: return None return fields[1 + fields.index('gi')] def get_gi_ids(filename): """Return a list of the GenInfo IDs of all sequences found in the file named filename""" with open(filename) as file: return [extract_gi_id(line) for line in file if line[0] == '>'] def get_gi_ids_from_files(filenames): """Return a list of the GenInfo IDs of all sequences found in the files whose names are contained in the collection filenames""" idlst = [] for filename in filenames: idlst += get_gi_ids(filename) return idlst
Search
Another common use of iterations is to search for an element that passes some
kind of test. This is not the same as a combine iteration—the result
of a combination is a property of all the elements of the collection,
whereas a search iteration is much like a search loop. Searching takes
many forms, not all of them iterations, but the one thing you’ll just
about always see is a return statement
that exits the function as soon as a matching element has been found.
If the end of the function is reached without finding a matching
element the function can end without explicitly returning a value,
since it returns None by default.
Suppose we have an enormous FASTA file and we need to extract from it a sequence with a specific GenBank ID. We don’t want to read every sequence from the file, because that could take much more time and memory than necessary. Instead, we want to read one entry at a time until we locate the target. This is a typical search. It’s also something that comprehensions can’t do: since they can’t incorporate statements, there’s no straightforward way for them to stop the iteration early.
As usual, we’ll build this out of several small functions. We’ll define four functions. The first is the “top-level” function; it calls the second, and the second calls the third and fourth. Here’s an outline showing the functions called by the top-level function:
search_FASTA_file_by_gi_id(id, filename)
FASTA_search_by_gi_id(id, fil)
extract_gi_id(line)
read_FASTA_sequence(fil)This opens the file and calls FASTA_search_by_gi_id to do the real work.
That function searches through the lines of the file looking for those
beginning with a '>'. Each time
it finds one it calls get_gi_id to
get the GenInfo ID from the line, if there is one. Then it compares
the extracted ID to the one it is looking for. If there’s a match, it
calls read_FASTA_sequence and
returns. If not, it continues looking for the next FASTA description
line. In turn, read_FASTA_sequence
reads and joins lines until it runs across a description line, at
which point it returns its result. Example 4-16 shows the
definition of the top-level function.
“Top-level” functions should almost always be very simple. They are entry points into the capabilities the other function definitions provide. Essentially, what they do is prepare the information received through their parameters for handling by the functions that do the actual work.
def search_FASTA_file_by_gi_id(id, filename): """Return the sequence with the GenInfo ID ID from the FASTA file named filename, reading one entry at a time until it is found""" id = str(id) # user might call with a number with open(filename) as file: return FASTA_search_by_gi_id(id, file)
Each of the other functions can be implemented in two ways. Both
FASTA_search_by_gi_id and read_FASTA_sequence can be implemented using
a loop or iteration. The simple function get_gi_id can be implemented with a
conditional expression or a conditional statement. Table 4-1 shows both
implementations for FASTA_search_by_gi_id.
The iterative implementation of FASTA_search_by_gi_id treats the file as a
collection of lines. It tests each line to see if it is the one that
contains the ID that is its target. When it finds the line it’s
seeking, it does something slightly different than what the template
suggests: instead of returning the line—the item found—it goes ahead
and reads the sequence that follows it.
Note
The templates in this book are not meant to restrict your code to specific forms: they are frameworks for you to build on, and you can vary the details as appropriate.
The next function—read_FASTA_sequence—shows another variation
of the search template. It too iterates over lines in the file—though
not all of them, since it is called after FASTA_search_by_gi_id has already read many
lines. Another way it varies from the template is that it accumulates
a string while looking for a line that begins with a '>'. When it finds one, it returns the
accumulated string. Its definition is shown in Table 4-2, and the
definition of get_gi_id is shown in
Table 4-3.
A special case of search iteration is where the result returned
is interpreted as a Boolean rather than the item found. Some search
iterations return False when a
match is found, while others return True. The exact form of the function
definition depends on which of those cases it implements. If finding a
match to the search criteria means the function should return False, then the last statement of the
function would have to return True
to show that all the items had been processed without finding a match.
On the other hand, if the function is meant to return True when it finds a match it is usually not
necessary to have a return at the
end, since None will be returned by
default, and None is interpreted as
false in logical expressions. (Occasionally, however, you really need
the function to return a Boolean value, in which case you would end
the function by returning False.)
Here are two functions that demonstrate the difference:
def rna_sequence_is_valid(seq): for base in seq: if base not in 'UCAGucag': return False return True def dna_sequence_contains_N(seq): for base in seq: if base == 'N': return True
Filter
Filtering is similar to searching, but instead of returning a result the first
time a match is found, it does something with each element for which
the match was successful. Filtering doesn’t stand on its own—it’s a
modification to one of the other kinds of iterations. This section
presents templates for some filter iterations. Each just adds a
conditional to one of the other templates. The condition is shown
simply as test item, but in practice that test could be
complex. There might even be a few initialization statements before
the conditional.
An obvious example of a Filtered Do is printing the header lines from a FASTA file. Example 4-17 shows how this would be implemented.
def print_FASTA_headers(filename): with open(filename) as file: for line in file: if line[0] == '>': print(line[1:-1])
As with Collect iterations in general, simple situations can be handled with comprehensions, while iterations can handle the more complex situations in which statements are all but unavoidable. For example, extracting and manipulating items from a file can often be handled by comprehensions, but if the number of items is large, each manipulation will create an unnecessarily large collection. Rather than collecting all the items and performing a sequence of operations on that collection, we can turn this inside out, performing the operations on one item and collecting only the result.
In many cases, once a line passes the test the function should
not return immediately. Instead, it should continue to read lines,
concatenating or collecting them, until the next time the test is
true. An example would be with FASTA-formatted files, where a function
might look for all sequence descriptions that contain a certain
string, then read all the lines of the sequences that follow them.
What’s tricky about this is that the test applies only to the lines
beginning with '>'. The lines of
a sequence do not provide any information to indicate whether they
should be included or not.
Really what we have here are two tests: there’s a preliminary
test that determines whether the primary test should be performed.
Neither applies to the lines that follow a description line in the
FASTA file, though. To solve this problem, we add a flag to govern the
iteration and set it by performing the primary test whenever the
preliminary test is true. Example 4-18 shows a function
that returns the sequence strings for all sequences whose descriptions
contain the argument string.
def extract_matching_sequences(filename, string): """From a FASTA file named filename, extract all sequences whose descriptions contain string""" sequences = [] seq = '' with open(filename) as file: for line in file: if line[0] == '>': if seq: # not first time through sequences.append(seq) seq = '' # next sequence detected includeflag = string in line # flag for later iterations else: if includeflag: seq += line[:-1] if seq: # last sequence in file is included sequences.append(seq) return sequences
The generalization of this code is shown in the following template.
A Filtered Combine is just like a regular Combine, except only elements that pass the test are used in the combining expression.
Example 4-13 showed a definition
for product. Suppose the collection
passed to product contained nonnumerical
elements. You might want the product function to skip nonnumerical values
instead of converting string representations of numbers to numbers.[24]
All that’s needed to skip nonnumerical values is a test that
checks whether the element is an integer or float and ignores it if it
is not. The function isinstance was
described briefly in Chapter 1; we’ll use that
here to check for numbers. Example 4-19 shows this new
definition for product.
def is_number(value): """Return True if value is an int or a float""" return isinstance(elt, int) or isinstance(elt, float) def product(coll): """Return the product of the numeric elements of coll""" result = 1.0 # initialize for elt in coll: if is_number(elt): result = result * float(elt) # combine element with accumulated result return result
What we’ve done here is replace the template’s
test with a call to is_number to perform the test. Suppose we
needed different tests at different times while computing the
product—we might want to ignore zeros or negative numbers, or we might
want to start at a different initial value (e.g., 1 if computing the product of only
integers). We might even have different actions to perform each time
around the iteration. We can implement many of these templates as
function definitions whose details are specified by parameters. Example 4-20 shows a completely general
combine function.
def combine(coll, initval, action, filter=None): """Starting at initval, perform action on each element of coll, finally returning the result. If filter is not None, only include elements for which filter(element) is true. action is a function of two arguments--the interim result and the element--which returns a new interim result.""" result = initval for elt in coll: if not filter or filter(elt): result = action(result, elt) return result
To add all the integers in a collection, we just have to call
combine with the right arguments:
combine(coll
0,
lambda result, elt: result + elt,
lambda elt: isinstance(elt, int)
)Nested iterations
One iteration often uses another. Example 4-21 shows a simple case—listing all the sequence IDs in files whose names are in a collection.
def list_sequences_in_files(filelist): """For each file whose name is contained in filelist, list the description of each sequence it contains""" for filename in filelist: print(filename) with open(filename) as file: for line in file: if line[0] == '>': print('\t', line[1:-1])
Nesting is not a question of physical containment of one piece
of code inside another. Following the earlier recommendation to write
short, single-purpose functions, Example 4-22 divides the previous
function, placing one iteration in each. This is still a nested
iteration, because the first function calls the second each time
around the for, and the second has
its own for statement.
def list_sequences_in_files(filelist): """For each file whose name is contained in filelist, list the description of each sequence it contains""" for filename in filelist: print(filename) with open(filename) as file: list_sequences_in_file(file) def list_sequences_in_file(file) for line in file: if line[0] == '>': print('\t', line[1:-1])
These examples do present nested iterations, but they don’t show what’s special about this kind of code. Many functions that iterate call other functions that also iterate. They in turn might call still other functions that iterate. Nested iterations are more significant when their “do something” parts involve doing something with a value from the outer iteration and a value from the inner iteration together.
Perhaps a batch of samples is to be submitted for sequencing with each of a set of primers:
for seq in sequences: for primer in primers: submit(seq, primer)
This submits a sequence and a primer for every combination of a
sequence from sequences and a primer from primers. In this case it doesn’t matter
which iteration is the outer and which is the inner, although if they
were switched the sequence/primer pairs would be submitted in a
different order.
Three-level iterations are occasionally useful—especially in bioinformatics programming, because codons consist of three bases. Example 4-23 shows a concise three-level iteration that prints out a simple form of the DNA codon table.
def print_codon_table():
"""Print the DNA codon table in a nice, but simple, arrangement"""
for base1 in DNA_bases: # horizontal section (or "group")
for base3 in DNA_bases: # line (or "row")
for base2 in DNA_bases: # vertical section (or "column")
# the base2 loop is inside the base3 loop!
print(base1+base2+base3,
translate_DNA_codon(base1+base2+base3),
end=' ')
print()
print()
>>> print_codon_table()
TTT Phe TCT Ser TAT Tyr TGT Cys
TTC Phe TCC Ser TAC Tyr TGC Cys
TTA Leu TCA Ser TAA --- TGA ---
TTG Leu TCG Ser TAG --- TGG Trp
CTT Leu CCT Pro CAT His CGT Arg
CTC Leu CCC Pro CAC His CGC Arg
CTA Leu CCA Pro CAA Gln CGA Arg
CTG Leu CCG Pro CAG Gln CGG Arg
ATT Ile ACT Thr AAT Asn AGT Ser
ATC Ile ACC Thr AAC Asn AGC Ser
ATA Ile ACA Thr AAA Lys AGA Arg
ATG Met ACG Thr AAG Lys AGG Arg
GTT Val GCT Ala GAT Asp GGT Gly
GTC Val GCC Ala GAC Asp GGC Gly
GTA Val GCA Ala GAA Glu GGA Gly
GTG Val GCG Ala GAG Glu GGG GlyRecursive iterations
Trees are an important class of data structure in computation: they provide the generality needed to represent branching information. Taxonomies and filesystems are good examples. A filesystem starts at the top-level directory of, say, a hard drive. That directory contains files and other directories, and those directories in turn contain files and other directories. The whole structure consists of just directories and files.
A data structure that can contain other instances of itself is said to be recursive. The study of recursive data structures and algorithms to process them is a major subject in computer science. Trees are the basis of some important algorithms in bioinformatics too, especially in the areas of searching and indexing.
While we won’t be considering such algorithms in this book, it
is important to know some rudimentary techniques for tree
representation and iteration. A simple while or for statement can’t by itself follow all the
branches of a tree. When it follows one branch, it may encounter
further branches, and at each juncture it can follow only one at a
time. It can only move on to the next branch after it’s fully explored
everything on the first one. In the meantime, it needs someplace to
record a collection of the remaining branches to be processed.
Each branch is just another tree. A function that processes a tree can call itself to process each of the tree’s branches. What stops this from continuing forever is that eventually subtrees are reached that have no branches; these are called leaves. A function that calls itself—or calls another function that eventually calls it—is called a recursive function.
Discussions of recursion are part of many programming texts and courses. It often appears mysterious until the idea becomes familiar, which can take some time and practice. One of the advantages of recursive functions is that they can express computations more concisely than other approaches, even when recursion isn’t actually necessary. Sometimes the code is so simple you can hardly figure out how it does its magic!
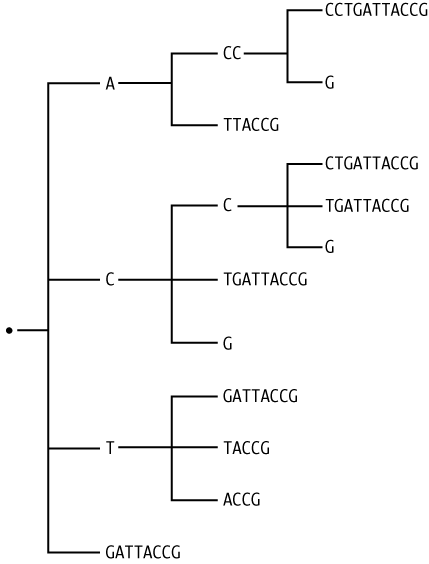
First, we’ll look at an example of one of the ways trees are used in bioinformatics. Some very powerful algorithms used in indexing and searching genomic sequences rely on what are called suffix trees. These are tree structures constructed so that every path from the root to a leaf produces a subsequence that is not the prefix of any other subsequence similarly obtained. The entire string from which the tree was constructed can be recovered by traversing all paths to leaf nodes, concatenating the strings encountered along the way, and collecting the strings obtained from each path. The longest string in the resulting collection is the original string. Figure 4-2 shows an example.
Algorithms have been developed for constructing and navigating such trees that do their work in an amount of time that is directly proportional to the length of the sequence. Normally algorithms dealing with tree-structured data require time proportional to N2 or at best N log N, where N is the length of the sequence. As N gets as large as is often required for genomic sequence searches, those quantities grow impossibly large. From this point of view the performance of suffix tree algorithms borders on the miraculous.
Our example will represent suffix trees as lists of lists of lists of... lists. The first element of each list will always be a string, and each of the rest of the elements is another list. The top level of the tree starts with an empty string. Example 4-24 shows an example hand-formatted to reflect the nested relationships.
['',
['A',
['CC',
['CCTGATTACCG'],
['G']
],
['TTACCG']
],
['C',
['C',
['CTGATTACCG'],
['TGATTACCG'],
['G']
],
['TGATTACCG'],
['G']
],
['T',
['GATTACCG'],
['TACCG'],
['ACCG']
],
['GATTACCG']
]Let’s assign tree1 to this
list and see what Python does with it. Example 4-25 shows an
ordinary interpreter printout of the nested lists.
['', ['A', ['CC', ['CCTGATTACCG'], ['G']], ['TTACCG']], ['C', ['C', ['CTGATTACCG'], ['TGATTACCG'], ['G']], ['TGATTACCG'], ['G']], ['T', ['GATTACCG'], ['TACCG'], ['ACCG']], ['GATTACCG']]
That output was one line, wrapped. Not very helpful. How much of
an improvement does pprint.pprint
offer?
>>> pprint.pprint(tree1)
['',
['A', ['CC', ['CCTGATTACCG'], ['G']], ['TTACCG']],
['C', ['C', ['CTGATTACCG'], ['TGATTACCG'], ['G']], ['TGATTACCG'], ['G']],
['T', ['GATTACCG'], ['TACCG'], ['ACCG']],
['GATTACCG']]This is a little better, since we can at least see the top-level structure. But what we want is output that approximates the tree shown in Figure 4-2. (We won’t go so far as to print symbols for lines and corners—we’re just looking to reflect the overall shape of the tree represented by the data structure.) Here’s the template for a recursive function to process a tree represented as just described here. (The information the tree contains could be anything, not just strings: whatever value is placed in the first position of the list representing a subtree is the value of that subtree’s root node.)
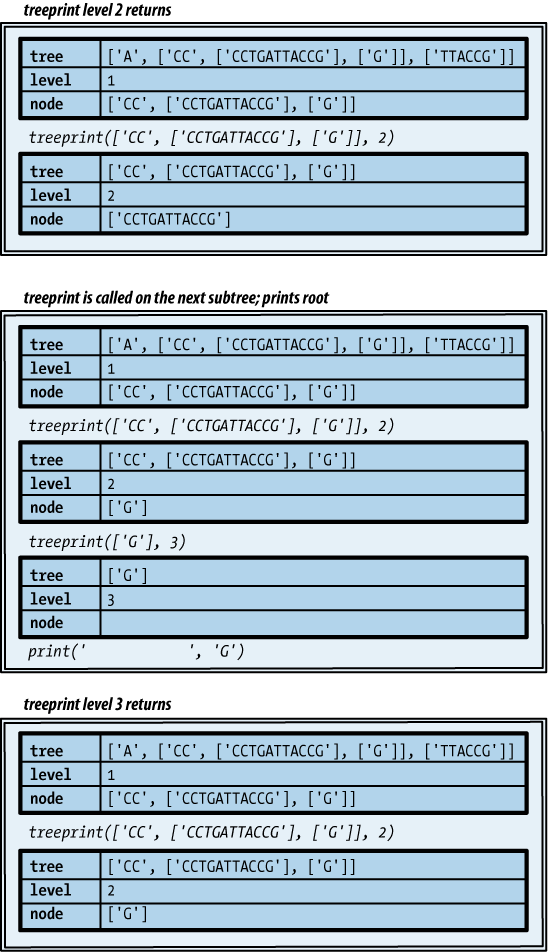
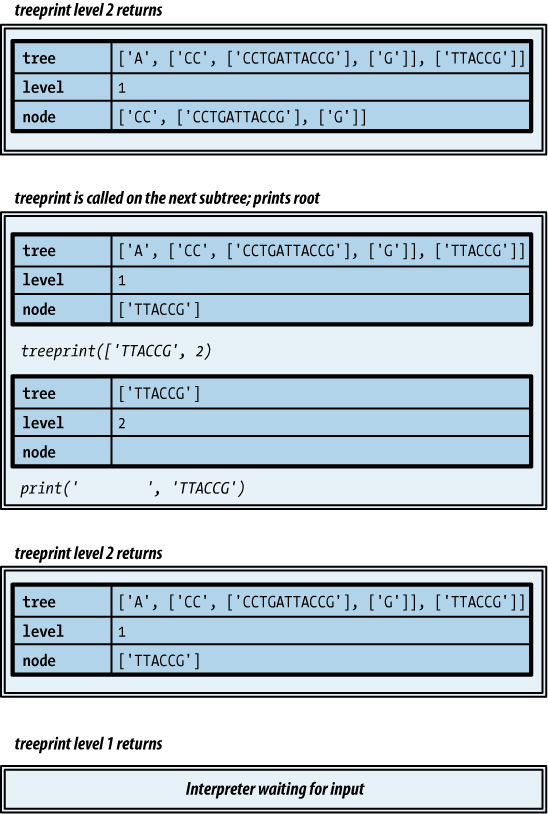
Do you find it difficult to believe that so simple a template can process a tree? Example 4-26 shows how it would be used to print our tree.
def treeprint(tree, level=0): print(' ' * 4 * level, tree[0], sep='') for node in tree[1:]: treeprint(node, level+1)
This produces the following output for the example tree. It’s not as nice as the diagram; not only are there no lines, but the root of each subtree is on a line before its subtrees, rather than centered among them. Still, it’s not bad for four lines of code!
A
CC
CCTGATTACCG
G
TTACCG
C
C
CTGATTACCG
TGATTACCG
G
TGATTACCG
G
T
GATTACCG
TACCG
ACCG
GATTACCGFigures 4-3, 4-4, and 4-5 illustrate the process that ensues
as the function in Example 4-26 does its work
with the list representing the subtree rooted at A.
Exception Handlers
Let’s return to Example 4-15, from our discussion of collection iteration. We’ll add a top-level function to drive the others and put all of the functions in one Python file called get_gi_ids.py. The contents of the file are shown in Example 4-27.
def extract_gi_id(description): """Given a FASTA file description line, return its GenInfo ID if it has one""" if line[0] != '>': return None fields = description[1:].split('|') if 'gi' not in fields: return None return fields[1 + fields.index('gi')] def get_gi_ids(filename): """Return a list of GenInfo IDs from the sequences in the FASTA file named filename""" with open(filename) as file: return [extract_gi_id(line) for line in file if line[0] == '>'] def get_gi_ids_from_files(filenames): """Return a list of GenInfo IDs from the sequences in the FASTA files whose names are in the collection filenames""" idlst = [] for filename in filenames: idlst += get_gi_ids(filename) return idlst def get_gi_ids_from_user_files(): response = input("Enter FASTA file names, separated by spaces: ") lst = get_gi_ids_from_files(response.split()) # assuming no spaces in file names lst.sort() print(lst) get_gi_ids_from_user_files()
We run the program from the command line, enter a few filenames, and get the results shown in Example 4-28.
%python get_gi_ids.pyEnter a list of FASTA filenames:aa1.fasta aa2.fasta aa3.fastaTraceback (most recent call last): File "get_gi_ids.py", line 27, in <module> get_gi_ids_from_user_files File "get_gi_ids.py", line 23, in get_gi_ids_from_user_files lst = get_gi_ids_from_files(files)) File "get_gi_ids.py", line 18, in get_gi_ids_from_files idlst += get_gi_ids(filename) File "get_gi_ids.py", line 10, in get_gi_ids with open(filename) as file: File "/usr/local/lib/python3.1/io.py", line 278, in __new__ return open(*args, **kwargs) File "/usr/local/lib/python3.1/io.py", line 222, in open closefd) File "/usr/local/lib/python3.1/io.py", line 619, in __init__ _fileio._FileIO.__init__(self, name, mode, closefd) IOError: [Errno 2] No such file or directory: 'aa2.fasta'
Python Errors
If you’ve executed any Python code you have written, you have probably already seen output like that in the previous example splattered across your interpreter or shell window. Now it’s time for a serious look at what this output signifies. It’s important to understand more than just the message on the final line and perhaps a recognizable filename and line number or two.
Tracebacks
As its first line indicates, the preceding output shows details of
pending functions. This display of information is called a traceback. There are two lines for
each entry. The first shows the name of the function that was called,
the path to the file in which it was defined, and the line number
where its definition begins, though not in that order. (As in this
case, you will often see <module> given as the module name on
the first line; this indicates that the function was called from the
top level of the file being run by Python or directly from the
interpreter.) The second line of each entry shows the text of the line
identified by the filename and line number of the first line, to save
you the trouble of going to the file to read it.
Note
Some of this will make sense to you now. Some of it won’t until you have more Python knowledge and experience. As the calls descend deeper into Python’s implementation, some technical details are revealed that we haven’t yet explored. It’s important that you resist the temptation to dismiss tracebacks as hopelessly complicated and useless. Even if you don’t understand all the details, tracebacks tell you very clearly what has happened, where, and, to some extent, why.
The problem causing the traceback in this example is clear
enough: the user included the file aa2.fasta in the list of files to be
processed, but when get_gi_id went
to open that file it couldn’t find it. As a result, Python reported an
IOError and stopped executing. It
didn’t even print the IDs that it had already found—it just
stopped.
Runtime errors
Is this what you want your program to do? You can’t prevent the user
from typing the name of a nonexistent file. While you could check that
each file exists before trying to open it (using methods from the
os module that we’ll be looking in
Chapter 6), this is only one of the many things
that could go wrong during the execution of your program. Maybe the
file exists but you don’t have read privileges for it, or it exists
but is empty, and you didn’t write your code to correctly handle that
case. Maybe the program encounters an empty line at the end of
the file and tries to extract pieces from it. Maybe the program tries
to compare incompatible values in an expression such as 4 < '5'.
By now you’ve probably encountered ValueError, TypeError, IndexError, IOError, and perhaps a few others. Each of
these errors is actually a type. Table 4-4 shows examples of common errors,
the type of error instance that gets created when they occur, and
examples of the messages that get printed.
Example | Error class | Message |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
| |
|
|
|
|
| |
[a] The second argument of [b] [c] Typing Ctrl-D on an empty line (Ctrl-Z on Windows)
ends input. Remember, though, that [d] Pressing Ctrl-C twice stops whatever Python is doing and returns to the interpreter. | ||
Even if get_gi_ids was
written to detect nonexistent files before trying to open them, what
should it do if it detects one? Should it just return None? Should it print its own error message
before returning None? If it
returns None, how can the function
that called it know whether that was because the file didn’t exist,
couldn’t be read, wasn’t a FASTA-formatted file, or just didn’t have
any sequences with IDs? If each function has to report to its caller
all the different problems it might have encountered, each caller will
have to execute a series of conditionals checking each of those
conditions before continuing with its own executions.
To manage this problem, languages provide exception handling mechanisms. These make it possible to ignore exceptions when writing most function definitions, while specifically designating, in relatively few places, what should happen when exceptions do occur. The term “exception” is used instead of “error” because if the program is prepared to handle a situation, it isn’t really an error when it arises. It becomes an error—an unhandled exception—if the program does not detect the situation. In that case, execution stops and Python prints a traceback with a message identifying the type of error and details about the problem encountered.
Exception Handling Statements
Python’s exception handling mechanism is implemented through the try statement. This looks and works much like
a conditional, except that the conditions are not tests you write, but
rather names of error classes.
The error class is one of the error names you’ll see printed out
on the last line of a traceback: IOError, ValueError, and so on. When a try statement begins, it starts executing the statements in the
try-statements block. If they complete
without any errors, the rest of the try statement is skipped and execution
continues at the next statement.
However, if an error of the type identified in the except clause occurs
during the execution of the try
block, something quite different happens: the call stack is “unwound” by
removing the calls to the functions “below” the one that contains the
try statement. Any of the
try-statements that haven’t yet executed are
abandoned. Execution continues with the statements in the except clause, and then moves on to the
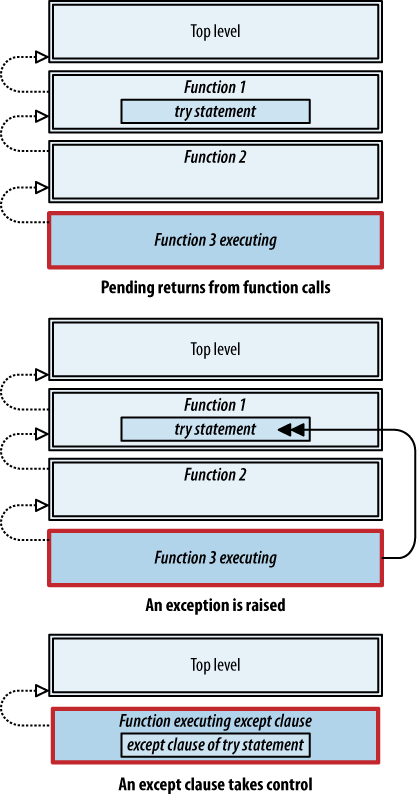
statement that follows the entire try/except statement. Figures 4-6
and 4-7 show the difference.
Optional features of exception handling statements
The try statement offers
quite a few options. The difficulty here is not so much in
comprehending all the details, although that does take some time. The
real challenge is to develop a concrete picture of how control “flows”
through function calls and Python’s various kinds of statements. Then
you can begin developing an understanding of the very different flow
induced by exceptions.
Now that we know how to handle errors, what changes might we
want to make in our little program for finding IDs? Suppose we’ve
decided we want the program to print an error message whenever it
fails to open a file, but then continue with the next one. This is
easily accomplished with one simple try statement:
def get_gi_ids(filename): try: with open(filename) as file: return [extract_gi_id(line) for line in file if line[0] == '>'] except IOError: print('File', filename, 'not found or not readable.') return []
Note that the except clause
returns an empty list rather than returning None or allowing the function to end without
a return (which amounts to the same
thing). This is because the function that calls this one will be
concatenating the result with a list it is accumulating, and since
None isn’t a sequence it can’t be
added to one. (That’s another TypeError you’ll often see, usually as
a result of forgetting to return a value from a function.) If you’ve
named the exception with as
name, you can print(name) instead of or in addition to your own message.
Incidentally, this with
statement:
with open('filename') as file:usefile
is roughly the same as:
try: file = open('filename')usefile finally: file.close()
The finally clause guarantees
that the file will be closed whether an error happens or not—and
without the programmer having to remember to close it. This is a great
convenience that avoids several kinds of common problems. The with statement requires only one line in
addition to the statements to be executed, rather than the four lines
required by the try version.
Exception handling and generator objects
An important special use of try
statements is with generator objects. Each call to next with a generator object as its argument
produces the generator’s next value. When there are no more values,
next returns the value of its
optional second argument, if one is provided. If not, a StopIteration error is raised.
There are two ways to use next: either you can provide a default value
and compare it to the value next
returns each time, or you can omit the argument and put the call to
next inside a try that has an except StopIteration clause. (An except clause with no exception class or a
finally would also catch the
error.)
An advantage of the exception approach is that the try statement that catches it can be several
function calls back; also, you don’t have to check the value returned
by next each time. This is
particularly useful when one function calls another that calls
another, and so on. A one-argument call to next in the innermost function and a
try statement in the top-level
function will terminate the entire process and hand control back to
the top-level function, which catches StopIteration.
Raising Exceptions
Exception raising isn’t limited to library functions—your code can raise them too.
The raise statement
The raise statement
is used to raise an exception and initiate exception
handling.
The exception-expression can be any
expression whose value is either an exception class or an instance of
one. If it is an exception class, the statement simply creates an
instance for you. Creating your own instance allows you to specify
arguments to the new instance—typically a message providing more
detail about the condition encountered. The class Exception can
be used for whatever purposes you want, and it can take an arbitrary
number of arguments. You can put statements like this in your
code:
raise Exception('File does not appear to be in FASTA format.', filename)The statements in any of a try statement’s
exception clauses can “reraise” an exception using a raise statement with no expression. In that
case, the stack unwinding resumes and continues until the next
try statement prepared to handle
the exception is encountered.
Not only can your code raise exceptions, but you can create your own exception classes and raise instances of those. (The next chapter shows you how to create your own classes.) It’s especially important for people building modules for other people to use, since code in a module has no way of knowing what code from the outside wants to do when various kinds of problems are encountered. The only reasonable thing to do is design modules to define appropriate exception classes and document them for users of the module so they know what exceptions their code should be prepared to handle.
Raising an exception to end a loop
The point was made earlier that exceptions aren’t necessarily errors. You
can use a combination of try and
raise statements as an alternative
way of ending loops. You would do this if you had written a long
sequence of functions that call each other, expecting certain kinds of
values in return. When something fails deep down in a sequence of
calls it can be very awkward to return None or some other failure value back
through the series of callers, as each of them would have to test the
value(s) it got back to see whether it should continue or itself
return None. A common example is
repeatedly using str.find in many
different functions to work through a large string.
Using exception handling, you can write code without all that
distracting error reporting and checking. Exceptional situations can
be handled by raising an error. The first function called can have a
“while-true” loop inside a try
statement. Whenever some function determines that nothing remains to
process, it can throw an exception. A good exception class for this
purpose is StopIteration,
which is used in the implementation of generators,
while-as statements, and other mechanisms
we’ve seen:
try: while(True):begin complicated multi-function input processingexcept StopIteration: pass... many definitions of functions that call each other; ...... wherever one detects the end of input, it does: ...raise StopIteration
Extended Examples
This section presents some extended examples that make use of the constructs described earlier in the chapter.
Extracting Information from an HTML File
Our first example in this section is based on the technique just discussed of raising an exception to end the processing of some text. Consider how you would go about extracting information from a complex HTML page. For example, go to NCBI’s Entrez Gene site (http://www.ncbi.nlm.nih.gov/sites/entrez), enter a gene name in the search field, click the search button, and then save the page as an HTML file. Our example uses the gene vWF.[25] Example 4-29 shows a program for extracting some information from the results returned. The patterns it uses are very specific to results saved from Entrez Gene, but the program would be quite useful if you needed to process many such pages.
endresults = '- - - - - - - - end Results - - - - - -'
patterns = ('</em>]',
'\n',
'</a></div><div class="rprtMainSec"><div class="summary">',
)
def get_field(contents, pattern, endpos):
endpos = contents.rfind(pattern, 0, endpos)
if endpos < 0:
raise StopIteration
startpos = contents.rfind('>', 0, endpos)
return (endpos, contents[startpos+1:endpos])
def get_next(contents, endpos):
fields = []
for pattern in patterns:
endpos, field = get_field(contents, pattern, endpos)
fields.append(field)
fields.reverse()
return endpos, fields
def get_gene_info(contents):
lst = []
endpos = contents.rfind(endresults, 0, len(contents))
try:
while(True):
endpos, fields = get_next(contents, endpos)
lst.append(fields)
except StopIteration:
pass
lst.reverse()
return lst
def get_gene_info_from_file(filename):
with open(filename) as file:
contents = file.read()
return get_gene_info(contents)
def show_gene_info_from_file(filename):
infolst = get_gene_info_from_file(filename)
for info in infolst:
print(info[0], info[1], info[2], sep='\n ')
if __name__ == '__main__':
show_gene_info_from_file(sys.argv[1]
if len(sys.argv) > 1
else 'EntrezGeneResults.html')Output for the first page of the Entrez Gene results for vWF looks like this:
Vwf
Von Willebrand factor homolog
Mus musculus
VWF
von Willebrand factor
Homo sapiens
VWF
von Willebrand factor
Canis lupus familiaris
Vwf
Von Willebrand factor homolog
Rattus norvegicus
VWF
von Willebrand factor
Bos taurus
VWF
von Willebrand factor
Pan troglodytes
VWF
von Willebrand factor
Macaca mulatta
vwf
von Willebrand factor
Danio rerio
VWF
von Willebrand factor
Gallus gallus
VWF
von Willebrand factor
Sus scrofa
Vwf
lectin
Bombyx mori
VWF
von Willebrand factor
Oryctolagus cuniculus
VWF
von Willebrand factor
Felis catus
VWF
von Willebrand factor
Monodelphis domestica
VWFL2
von Willebrand Factor like 2
Ciona intestinalis
ADAMTS13
ADAM metallopeptidase with thrombospondin type 1 motif, 13
Homo sapiens
MADE_03506
Secreted protein, containing von Willebrand factor (vWF) type A domain
Alteromonas macleodii 'Deep ecotype'
NOR51B_705
putative secreted protein, containing von Willebrand factor (vWF) type A domain
gamma proteobacterium NOR51-B
BLD_1637
von Willebrand factor (vWF) domain containing protein
Bifidobacterium longum DJO10A
NOR53_416
secreted protein, containing von Willebrand factor (vWF) type A domain
gamma proteobacterium NOR5-3This code was developed in stages. The first version of the
program had separate functions get_symbol, get_name, and get_species. Once they were cleaned up and
working correctly it became obvious that they each did the same thing,
just with a different pattern. They were therefore replaced with a
single function that had an additional parameter for the search
pattern.
The original definition of get_next contained repetitious lines. This
definition replaces those with an iteration over a list of patterns.
These changes made the whole program easily extensible. To extract more
fields, we just have to add appropriate search patterns to the patterns list.
It should also be noted that because the second line of some
entries showed an “Official Symbol” and “Name” but others didn’t, it
turned out to be easier to search backward from the end of the file. The
first step is to find the line demarcating the end of the results. Then
the file contents are searched in reverse for each pattern in turn, from
the beginning of the file to where the last search left off. (Note that
although you might expect it to be the other way around, the arguments
to rfind are interpreted just like
the arguments to find, with the
second less than the third.)
The Grand Unified Bioinformatics File Parser
This section explores some ways the process of reading information from text files can be generalized.
Reading the sequences in a FASTA file
Example 4-30 presents
a set of functions for reading the sequences in a FASTA file. They are actually quite general, and can
work for a variety of the kinds of formats typically seen in
bioinformatics. The code is a lot like what we’ve seen in earlier
examples. All that is needed to make these functions work for a
specific file format is an appropriate definition of skip_intro and next_item.
def get_items_from_file(filename, testfn=None): """Return all the items in the file named filename; if testfn then include only those items for which testfn is true""" with open(filename) as file: return get_items(file, testfn) def find_item_in_file(filename, testfn=None): """Return the first item in the file named filename; if testfn then return the first item for which testfn is true""" with open(filename) as file: return find_item(file, testfn) def find_item(src, testfn): """Return the first item in src; if testfn then return the first item for which testfn is true""" gen = item_generator(src, testfn) item = next(gen) if not testfn: return item else: try: while not testfn(item): item = next(gen) return item except StopIteration: return None def get_items(src, testfn=None): """Return all the items in src; if testfn then include only those items for which testfn is true""" return [item for item in item_generator(src) if not testfn or testfn(item)] def item_generator(src): """Return a generator that produces a FASTA sequence from src each time it is called""" skip_intro(src) seq = '' description = src.readline().split('|') line = src.readline() while line: while line and line[0] != '>': seq += line line = src.readline() yield (description, seq) seq = '' description = line.split('|') line = src.readline() def skip_intro(src): """Skip introductory text that appears in src before the first item""" pass # no introduction in a FASTA file
The functions get_items_from_file and find_item_in_file simply take a filename and
call get_items and find_item, respectively. If you already have
an open file, you can pass it directly to get_items or find_item. All four functions take an
optional filter function. If one is provided, only items for which the
function returns true are included. Typically, a filter function like
this would be a lambda expression. Note that find_item can be called repeatedly on the
same open file, returning the next item for which testfn is true, because after the first one
is found the rest of the source is still available for reading.
next_item is a generator
version of the functions we’ve seen for reading FASTA entries. It
reads one entry each time it is called, returning the split
description line and the sequence as a pair. This function and
possibly skip_intro would need to
be defined differently for different file formats. The other four
functions stay the same.
Generalized parsing
Extracting a structured representation from a text file is known as parsing. Python, for example, parses text typed at the interpreter prompt or imported from a module in order to convert it into an executable representation according to the language’s rules. Much of bioinformatics programming involves parsing files in a wide variety of formats. Despite the ways that formats differ, programs to parse them have a substantial underlying similarity, as reflected in the following template.
Parsing GenBank Files
Next, we’ll look at an example of applying the generalized parser template to read features and sequences from GenBank flat files.[26] There are many ways to navigate in a browser to get a page in GenBank format from the NCBI website.[27] For instance, if you know the GenInfo Identifier (GI), you can get to the corresponding GenBank record using the URL http://www.ncbi.nlm.nih.gov/nuccore/ followed by the GI number. Then, to download the page as a flat text file, simply click on the “Download” drop-down on the right side of the page just above the name of the sequence and select “GenBank” as the format. The file will be downloaded as sequence.gb to your browser’s default download directory.
There’s a great deal of information in these GenBank entries. For
this example we just want to extract the accession code, GI number,
feature information, and sequence. Example 4-31 shows the code needed
to implement the format-specific part of the unified parser template:
skip_intro and next_item. For a given format, the
implementation of either of these two functions may require other
supporting functions.
def get_GenBank_items_and_sequence_from_file(filename): with open(filename) as file: return [get_ids(file), get_items(file), get_sequence(file)] def get_ids(src): line = src.readline() while not line.startswith('VERSION'): line = src.readline() parts = line.split() # split at whitespace; removes \n assert 3 == len(parts), parts # should be VERSION acc GI:id giparts = parts[2].partition(':') assert giparts[2], giparts # if no colon, [1] & [2] are empty assert giparts[2].isdigit() # all numbers? return (parts[1], giparts[2]) def get_sequence(src): """Return the DNA sequence found at end of src""" # When this is called the ORIGIN line should have just been read, # so we just have to read the sequence lines until the // at the end seq = '' line = src.readline() while not line.startswith('//'): seq += line[10:-1].replace(' ', '') line = src.readline() return seq def skip_intro(src): """Skip introductory text that appears before the first item in src""" line = src.readline() while not line.startswith('FEATURES'): line = src.readline() attribute_prefix = 21*' ' + '/' def is_attribute_start(line): return line and line.startswith(attribute_prefix) def is_feature_start(line): return line and line[5] != ' ' def next_item(src): """Return a generator that produces a FASTA sequence from src each time it is called""" skip_intro(src) line = src.readline() while not line.startswith('ORIGIN'): assert is_feature_start(line) # line should start a feature feature, line = read_feature(src, line) # need to keep line to feed back to read_feature yield feature def read_feature(src, line): feature = line.split() props = {} line = src.readline() while not is_feature_start(line): key, value = line.strip()[1:].split('=') # remove initial / and split into [feature, value] if value[0] == '"': value = value[1:] # remove first "; remove final " later fullvalue, line = read_value(src, line, value) # need to keep line to feed back to read_value props[key] = fullvalue feature.append(props) return feature, line def read_value(src, line, value): line = src.readline() while (not is_attribute_start(line) and not is_feature_start(line)): value += line.strip() line = src.readline() if value[-1] == '"': value = value[:-1] # remove final " return value, line
The template is meant as a helpful outline, not a restrictive structure. A program written according to this template may add its own actions. In this case, the “items” to be read are the features. Before reading the features, the program extracts the accession and GI numbers. After all the features have been read, an extra step is needed to read the sequence. The top-level function returns those items in a list: a pair containing the accession and GI numbers, the list of features, and the sequence. Each feature is a list containing the type of the feature, the range of bases it covers, and a dictionary of key/value pairs defining properties.
For the GenBank sample record saved from http://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html, the result of executing the code would be as follows (sequence strings have been truncated to fit on a single line, and explanations have been added to the output as comments):
>>> data = get_genbank_items_and_sequence_from_file('sequences/sample.gb')
>>> pprint.pprint(data)
[('U49845.1', '1293613'), # (accession, GI)
[['source', # first feature is source
'1..5028', # range of source within base seq
{'chromosome': 'IX',
'db_xref': 'taxon:4932', # reference to Taxonomy DB
'map': '9',
'organism': 'Saccharomyces cerevisiae'}],
['CDS', # coding sequence
'<1..206', # seq from base 1 to 206, 5' partial
{'codon_start': '3', # translation starts at 3
'db_xref': 'GI:1293614', # protein GI
'product': 'TCP1-beta', # protein produced by the CDS
'protein_id': 'AAA98665.1', # protein accession
'translation': 'SSIYNGIS...'}], # amino acid sequence
# gene AXL2 spans nucleotides 687 through 3158
['gene', '687..3158', {'gene': 'AXL2'}],
['CDS',
'687..3158', # a complete coding sequence
{'codon_start': '1',
'db_xref': 'GI:1293615',
'function': 'required for axial budding pattern of S.cerevisiae',
'gene': 'AXL2',
'note': 'plasma membrane glycoprotein',
'product': 'Axl2p',
'protein_id': 'AAA98666.1',
'translation': 'MTQLQISL...'}],
['gene', 'complement(3300..4037)', {'gene': 'REV7'}],
['CDS',
'complement(3300..4037)', # CDS is on opposite strand
{'codon_start': '1',
'db_xref': 'GI:1293616',
'gene': 'REV7',
'product': 'Rev7p',
'protein_id': 'AAA98667.1',
'translation': 'MNRWVEKW...'}]],
# base sequence:
'gatcctccatatacaacggtatctccacctcaggtttagatctcaacaacggaaccattgcc...']Translating RNA Sequences
Next, we’re going to build a program to translate RNA sequences directly into strings of three-letter amino acid abbreviations. We actually got a start on this in the previous chapter, where we defined a codon table and a lookup function. They will be repeated here for convenience.
Step 1
Example 4-32 begins a series of function definitions interleaved with brief explanatory text and sample printouts.
RNA_codon_table = {
# Second Base
# U C A G
# U
'UUU': 'Phe', 'UCU': 'Ser', 'UAU': 'Tyr', 'UGU': 'Cys', # UxU
'UUC': 'Phe', 'UCC': 'Ser', 'UAC': 'Tyr', 'UGC': 'Cys', # UxC
'UUA': 'Leu', 'UCA': 'Ser', 'UAA': '---', 'UGA': '---', # UxA
'UUG': 'Leu', 'UCG': 'Ser', 'UAG': '---', 'UGG': 'Trp', # UxG
# C
'CUU': 'Leu', 'CCU': 'Pro', 'CAU': 'His', 'CGU': 'Arg', # CxU
'CUC': 'Leu', 'CCC': 'Pro', 'CAC': 'His', 'CGC': 'Arg', # CxC
'CUA': 'Leu', 'CCA': 'Pro', 'CAA': 'Gln', 'CGA': 'Arg', # CxA
'CUG': 'Leu', 'CCG': 'Pro', 'CAG': 'Gln', 'CGG': 'Arg', # CxG
# A
'AUU': 'Ile', 'ACU': 'Thr', 'AAU': 'Asn', 'AGU': 'Ser', # AxU
'AUC': 'Ile', 'ACC': 'Thr', 'AAC': 'Asn', 'AGC': 'Ser', # AxC
'AUA': 'Ile', 'ACA': 'Thr', 'AAA': 'Lys', 'AGA': 'Arg', # AxA
'AUG': 'Met', 'ACG': 'Thr', 'AAG': 'Lys', 'AGG': 'Arg', # AxG
# G
'GUU': 'Val', 'GCU': 'Ala', 'GAU': 'Asp', 'GGU': 'Gly', # GxU
'GUC': 'Val', 'GCC': 'Ala', 'GAC': 'Asp', 'GGC': 'Gly', # GxC
'GUA': 'Val', 'GCA': 'Ala', 'GAA': 'Glu', 'GGA': 'Gly', # GxA
'GUG': 'Val', 'GCG': 'Ala', 'GAG': 'Glu', 'GGG': 'Gly' # GxG
}
def translate_RNA_codon(codon):
return RNA_codon_table[codon]Step 2
The next step is to write a function that translates an RNA base
string into a string of the corresponding three-letter amino acid
abbreviations. The optional step argument
to range is useful for this.
Testing with assertions while this code was being developed revealed
the need to ignore the last base or two of sequences whose length is
not a multiple of 3, something not considered when the code was first
written. The expression len(seq)%3
gives the remainder when the length of the sequence is divided by 3—we
have to subtract that from len(seq)
so we don’t try to process an excess base or two at the end of the
sequence. The new example is shown in Example 4-33.
Step 3
Next, we take care of frame shifts and add printing functions with the functions shown in Example 4-34.
def translate_in_frame(seq, framenum): """Return the translation of seq in framenum 1, 2, or 3""" return translate(seq[framenum-1:]) def print_translation_in_frame(seq, framenum, prefix): """Print the translation of seq in framenum preceded by prefix""" print(prefix, framenum, ' ' * framenum, translate_in_frame(seq, framenum), sep='') def print_translations(seq, prefix=''): """Print the translations of seq in all three reading frames, each preceded by prefix""" print('\n' ,' ' * (len(prefix) + 2), seq, sep='') for framenum in range(1,4): print_translation_in_frame(seq, framenum, prefix) >>>print_translations('ATGCGTGAGGCTCTCAA')ATGCGTGAGGCTCTCAA 1 MetArgGluAlaLeu 2 CysValArgLeuSer 3 Ala---GlySerGln >>>print_translations('ATGATATGGAGGAGGTAGCCGCGCGCCATGCGCGCTATATTTTGGTAT')ATGATATGGAGGAGGTAGCCGCGCGCCATGCGCGCTATATTTTGGTAT 1 MetIleTrpArgArg---ProArgAlaMetArgAlaIlePheTrpTyr 2 ---TyrGlyGlyGlySerArgAlaProCysAlaLeuTyrPheGly 3 AspMetGluGluValAlaAlaArgHisAlaArgTyrIleLeuVal
Step 4
Now we are ready to find the open reading frames. (We make the
simplifying assumption that
we’re using the standard genetic code.) The second and third functions
here are essentially the same as
in the previous step, except that they call translate_with_open_reading_frames
instead of translate_in_frame.
Example 4-35 shows the new
definitions.
def translate_with_open_reading_frames(seq, framenum): """Return the translation of seq in framenum (1, 2, or 3), with ---'s when not within an open reading frame; assume the read is not in an open frame when at the beginning of seq""" open = False translation = "" seqlength = len(seq) - (framenum - 1) for n in range(frame-1, seqlength - (seqlength % 3), 3): codon = translate_RNA_codon(seq[n:n+3]) open = (open or codon == "Met") and not (codon == "---") translation += codon if open else "---" return translation def print_translation_with_open_reading_frame(seq, framenum, prefix): print(prefix, framenum, ' ' * framenum, translate_with_open_reading_frames(seq, framenum), sep='') def print_translations_with_open_reading_frames(seq, prefix=''): print('\n', ' ' * (len(prefix) + 2), seq, sep='') for frame in range(1,4): print_translation_with_open_reading_frame(seq, frame, prefix) >>>print_translations('ATGCGTGAGGCTCTCAA')ATGCGTGAGGCTCTCAA 1 MetArgGluAlaLeu 2 --------------- 3 --------------- >>>print_translations('ATGATATGGAGGAGGTAGCCGCGCGCCATGCGCGCTATATTTTGGTAT')ATGATATGGAGGAGGTAGCCGCGCGCCATGCGCGCTATATTTTGGTAT 1 MetIleTrpArgArg------------MetArgAlaIlePheTrpTyr 2 --------------------------------------------- 3 ---MetGluGluValAlaAlaArgHisAlaArgTyrIleLeuVal
Step 5
Finally, we print the sequence both forward and backward.
Getting the reverse of a sequence is easy, even though there’s no
function for it: seq[::-1].
Remember that trick, as you will need it any time you want to reverse
a string. Working with biological sequence data, that will be quite
often! Example 4-36 shows the
final piece of the code.
def print_translations_in_frames_in_both_directions(seq): print_translations(seq, 'FRF') print_translations(seq[::-1], 'RRF') def print_translations_with_open_reading_frames_in_both_directions(seq): print_translations_with_open_reading_frames(seq, 'FRF') print_translations_with_open_reading_frames(seq[::-1], 'RRF') >>>print_translations('ATGCGTGAGGCTCTCAA')ATGCGTGAGGCTCTCAA FRF1 MetArgGluAlaLeu FRF2 --------------- FRF3 --------------- AACTCTCGGAGTGCGTA RRF1 --------------- RRF2 --------------- RRF3 --------------- >>>print_translations('ATGATATGGAGGAGGTAGCCGCGCGCCATGCGCGCTATATTTTGGTAT')ATGATATGGAGGAGGTAGCCGCGCGCCATGCGCGCTATATTTTGGTAT FRF1 MetIleTrpArgArg------------MetArgAlaIlePheTrpTyr FRF2 --------------------------------------------- FRF3 ---MetGluGluValAlaAlaArgHisAlaArgTyrIleLeuVal TATGGTTTTATATCGCGCGTACCGCGCGCCGATGGAGGAGGTATAGTA RRF1 ------------------------------------------------ RRF2 MetValLeuTyrArgAlaTyrArgAlaProMetGluGluVal--- RRF3 ---------------------------------------------
Constructing a Table from a Text File
Our next project will be to construct a table from a text file. We’ll use the file located at http://rebase.neb.com/rebase/link_bionet, which contains a list of restriction enzymes and their cut sites. The file has a simple format.[28] Each line of data has the form:
EnzymeName (Prototype) ... spaces ... CutSite
A “prototype” in this data set is the first enzyme to be discovered with the specified cut site. Lines that represent prototypes do not have anything in the “Prototype” column.
Several lines of information appear at the beginning of the file,
before the actual data. To ensure that our program ignores these lines
we’ll use one of our usual skip_intro-type functions, here called
get_first_line. A look at the file
shows that the first line of data is the first one that begins with an
A. This is certainly not an acceptable approach for “production”
software, since the organization of the file might change in the future,
but it’s good enough for this example. The end of the file may have some
blank lines, and we’ll need to ignore those too.
To represent the data in this file we’ll construct a dictionary whose keys are enzyme names and whose values are the cut sites. We’ll make this simple and ignore the information about prototypes. Because there are so many details we are going to take things a step at a time. This is how you should work on your programs too.
Step 1
The general outline of the program will be:
Initialize the enzyme table.
Skip introductory lines, returning the first real line.
While line is not empty:
Parse line.
Store entry in the enzyme table.
Read another line.
Turning those steps into function names and adding as few
details as we can get away with, we write some essentially empty
functions (see Example 4-37). For
example, get_first_line just
returns an empty string; in a sense it’s done its job, which is to
return a line.
def load_enzyme_table(): return load_enzyme_data_into_table({}) # start with empty dictionary def load_enzyme_data_into_table(table): line = get_first_line() while not end_of_data(line): parse(line) store_entry(table) line = get_next_line() return table def get_first_line(): return '' # stop immediately def get_next_line(): return ' ' # so it stops after getting the first line def end_of_data(line): return True def parse(line): return line def store_entry(table): pass # testing: def test(): table = load_enzyme_table() assert len(table) == 0 print('All tests passed.') test()
Step 2
We can fill in the details of some of these functions
immediately. Do not be disturbed that some of the definitions remain
trivial even after all the changes we’ll make. We might want to modify
this program to use with a different file format, and there’s no
guarantee that get_next_line, for
instance, will always be as simple as it is here. Using all these
function names makes it very clear what the code is doing without
having to comment it.
In the following steps, changes from and additions to the previous step are highlighted. They include:
Binding names to the result of
parse(using tuple unpacking)Passing the key and value obtained from
parsetostore_entryReturning a result from
first_lineto use in testingImplementing
end_of_dataSplitting the line into fields and returning the first and last using tuple packing
Implementing
store_entry
Many of the functions continue to have “pretend” implementations in step 2, which is shown in Example 4-38.
def load_enzyme_table():
return load_enzyme_data_into_table({})
# start with empty dictionary
def load_enzyme_data_into_table(table)
line = get_first_line()
while not end_of_data(line):
key, value = parse(line)
store_entry(table, key, value)
line = get_next_line()
return table
def get_first_line():
return 'enzymeA (protoA) CCCGGG'
# return a typical line
def get_next_line():
return ' ' # so it stops after getting the first line
def end_of_data(line):
return len(line) < 2
# 0 means end of file, 1 would be a blank line
def parse(line):
fields = line.split()
# with no argument, split splits at whitespace
# tuple packing (omitting optional parens)
return fields[0], fields[-1]
# avoiding having to determine whether there are 2 or 3
def store_entry(table, key, value):
table[key] = value
def test():
table = load_enzyme_table()
assert len(table) == 1
result = parse('enzymeA (protoA) CCCGGG')
assert result == ('enzymeA', 'CCCGGG'), result
print('All tests passed.')
test()Step 3
In the next step, we actually read from the file. It’s silly to try to wrestle with a large datafile while you are writing the functions to handle it. Instead, extract a small bit of the actual file and put it in a “test file” that you can use until the program seems to work. We’ll construct a file called rebase_test_data01.txt that contains exactly the following text:
some introductory text more introductory text AnEnzyme (APrototype) cutsite1 APrototype cutsite2
Changes in this step include making some of the definitions more realistic:
Adding a filename parameter to
load_enzyme_tableEmbedding most of that function in a loop
Adding a call to
printfor debugging purposesPassing the open file object to
return_first_lineandget_next_lineImplementing
get_first_lineandget_next_line
Example 4-39 illustrates the third step.
def load_enzyme_table(data_filename): with open(data_filename) as datafile: return load_enzyme_data_into_table(datafile, {}) def load_enzyme_data_into_table(datafile, table) line = get_first_line(datafile) while not end_of_data(line): print(line, end='') key, value = parse(line) store_entry(table, key, value) line = get_next_line(datafile) return table def get_first_line(fil): line = fil.readline() while line and not line[0] == 'A': line = fil.readline() return line def get_next_line(fil): return fil.readline() def end_of_data(line): return len(line) < 2 def parse(line): fields = line.split() return fields[0], fields[-1] def store_entry(table, key, value): table[key] = value def test(): print() datafilename = 'rebase_test_data01.txt' table = load_enzyme_table(datafilename) assert len(table) == 2, table result = parse('enzymeA (protoA) CCCGGG') assert result == ('enzymeA', 'CCCGGG'), result print() print('All tests passed.') test()
Step 4
Finally, we clean up some of the code (not shown here), use the real file, and test some results. Example 4-40 shows step 4.
# This step uses the definitions of the previous step unchanged, except # that the call to print in load_enzyme_data_into_table could be removed def test(): print() datafilename = 'link_bionet.txt' table = load_enzyme_table(datafilename) # check first entry from file: assert table['AaaI'] == 'C^GGCCG' # check an ordinary entry with a prototype: assert table['AbaI'] == 'T^GATCA', table # check an ordinary entry that is a prototype: assert table['BclI'] == 'T^GATCA', table # check last entry from file: assert table['Zsp2I'] == 'ATGCA^T' assert len(table) == 3559, len(table) print() print('All tests passed.')
Step 5
If we wanted to, we could also add a function for printing the table in a simpler format to make it easier to read in the future and a corresponding function for doing that reading. We can print each entry of the table on a separate line with the name of the enzyme separated by a tab from the sequence it recognizes. Example 4-41 shows these two simple functions.
def write_table_to_filename(table, data_filename): """Write table in a simple format to a file named data_filename""" with open(data_filename, 'w') as file: write_table_entries(table, files) def write_table_entries(table, datafile): for enzyme in sorted(table.keys()): print(enzyme, table[enzyme], sep=' ', file=datafile) def read_table_from_filename(data_filename): """Return a table read from the file named data_filename that was previously written by write_table_to_filename""" with open(data_filename) as file: return read_table_entries(file, {}) def read_table_entries(datafile): for line in datafile: fields = line.split() table[fields[0]] = fields[1] return table
Tips, Traps, and Tracebacks
Tips
Prefer comprehensions to loops or iterations that collect values.
Look for opportunities to use conditional expressions—
(val1 if val2 else val3)—instead ofif/elsestatements when the two clauses contain just an expression or assign the same name.The mechanisms discussed in this chapter are the core of Python programs. You should review the chapter from time to time to better understand parts you didn’t completely follow the first time through.
Use the templates as a reference—they capture a large portion of the ways control statements are used.
In general, if a function is to return
Trueif a certain condition holds andFalseotherwise, spelling out the following is technically “silly”:if
condition: return True else: return FalseInstead, just write
returncondition. For example, instead of:if len(seq1) > len(seq2): return True else: return False
write:
return len(seq1) > len(seq2)The result of the comparison is
TrueorFalse.When it is last in the function, a statement such as the following (without an
elseclause):if
boolean-condition: returnexpressioncan be more concisely written as:
return
boolean-conditionandexpressionThis expression will return
Falseifconditionis false and the value ofexpressionif it is true. The primary reason you would need to use the conditional statement instead ofexpressionis if you specifically want the function to returnNoneifconditionis false, rather thanFalse.While assertions are valuable for testing code and for checking conditions at certain points in function definitions while the definitions are still under development, do not use assertions as a coding technique in place of conditionals. For example, if you want to check the validity of some input, do that in a conditional statement and take appropriate action if the condition is violated, such as raising an error. Do not simply assert that condition.[29]
The first Python code in a file should be a docstring. If enclosed in triple single or double quotes, the docstring can be as many lines long as you want. If your file is imported (rather than executed), the module that will be created for it will store that docstring, and Python’s help facility will be able to display it. You’ll probably want to comment the file to explain what it is, even if only for your own future reference, so you might as well get in the habit of using docstrings rather than comments.
You should make a choice about how you will use single single, single double, triple single, and triple double quotes, then follow that convention in a consistent way. The reason for using each of the four kinds of quotes in a different way is that it makes it easier to search for appearances in your code of a string that serves a particular purpose, such as a docstring.
The following choices were made for the code in this book and the downloadable code files:
- Single single quotes
Used for short strings
- Single double quotes
Used for short strings with internal single quotes
- Triple double quotes
Used for docstrings
- Triple single quotes
Used for long value strings
The difference between single single quotes and single double quotes is not so important, but it’s better to use one most of the time. Don’t forget that you can include a single quote inside a single-quoted string (or a double quote inside a double-quoted string) simply by putting a backslash before it.
If a compilation error points to a line that appears to be OK, the problem is probably on the previous line. Check that all “structural” lines of compound statements—
def,if,else,elif,while,for,try,except, andfinally—end with a colon.When your IDE indents a line to an unexpected level, don’t fight it or ignore it—it’s giving you very useful information about a problem in your code. Make sure that each colon in a compound statement is at the end of its “logical line” (i.e., that the beginning and any continuation lines are treated as one). The only keywords that are followed by a colon (and therefore nothing else on that line) are
else,try, andfinally.Here’s a useful debugging function. It is better than just using
assertfor testing code because an assertion failure stops the program, whereas failures identified by calls to this function do not:def expect_equal(expected, result): """Return expected == result, printing an error message if not true; use by itself or with assert in a statement such as: assert expect_equal(3438, count_hypothetical_proteins(gbk_filename)""" if expected == result: return True print('Expected', expected, 'but got', result)
Traps
Many functions that collect values, whether using a loop or an iteration, must avoid adding to the collection on the first pass through the loop or iteration. When that is the case, the loop or iteration must be followed by a statement that adds the last item to the collection.
When processing data read from outside the program, do not trust that all of it is in the expected form. There may be missing or aberrant lines or entries; for example, an enzyme cut site that should be
?to mean “unknown” may be the digit7instead.It is rarely a good idea to process large files by calling
readlines. Rather, process one line at a time, by callingreadlinein a loop, by using a comprehension over the lines of the file, or by using aforstatement. This avoids creating enormous lists that occupy large amounts of memory.
Tracebacks
Following are some representative error messages:
AttributeError: 'range' object has no attribute 'index'EOFErrorIndentationError: unexpected indentThis can happen (well) after a
trystatement with noexceptorfinallyclause.IOError: [Errno 2] No such file or directory: 'aa2.fasta'There are a number of
IOErrorvariations, each with a differentErrnoand message format.KeyboardInterruptKeyErrorAn attempt has been made to reference a dictionary element by a key not present in the dictionary.
TypeError: unorderable types int() < str()An attempt has been made to compare two values of different types.
TypeError: 'NoneType' object is not iterableThe code contains a
forstatement that is iterating over something that has no value. This is not the same as iterating over an empty collection, which, although it does nothing, does not cause an error. Errors that mentionNoneTypeare almost always caused by a function meant to return a value that does not include areturnstatement.object is not iterable: 'builtin_function_or_method'A function name was used in place of a function call in the
inpart of a comprehension orforstatement; i.e., you forgot the parentheses. This is a common mistake when callingdict.keys(),dict.values(), anddict.items().
[20] The with statement is more
general than how it was described in Chapter 2: it actually does
not need to name the object of the with in an as portion of the statement, and its use is
not limited to files. However, the way it was described is the only way
it is used in this book.
[21] Strictly speaking, a function call expression (not a function definition) also affects the flow of control since it causes execution to proceed in the body of the function. Function calls can appear as separate statements or as part of expressions; either way, execution of the statements that follow or the rest of the expression is suspended until the function returns. From the point of view of the code calling the function, it is a single step that does not affect the order in which statements are executed.
[22] The len function is a bit
different: while it could be implemented by counting the elements one
at a time, most types implement length more directly.
[23] The term “reduce” comes from the mathematical idea that a one-dimensional collection is reduced to a “zero”-dimensional “scalar” value.
[24] Spreadsheet applications, for example, typically skip nonnumbers when performing numeric operations like “sum” on a row or column, rather than producing an error.
[25] vWF stands for “von Willebrand Factor,” which plays a role in von Willebrand disease, the most common human hereditary coagulation abnormality. There are several forms of the disease, other genes involved, and complex hereditary patterns.
[26] See http://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html for an example and an explanation.
[27] See http://www.ncbi.nlm.nih.gov.
[28] The site contains files in other formats with more information. We’ll use one of those later in the book.
[29] Other than coding style, the issue here is that assertions
are meant to be executed only during development, not in a
“production” version of a program. When running Python from the
command line, the -O option
can be added to optimize a few aspects of the execution, one of
which is to ignore assertions.
Get Bioinformatics Programming Using Python now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.